AMD’s Zen 5 is a missed opportunity in messaging
Last week AMD did their 'Tech Day' and it was anything but.
Read more ▶
The post AMD’s Zen 5 is a missed opportunity in messaging appeared first on SemiAccurate.
Last week AMD did their 'Tech Day' and it was anything but.
Read more ▶
The post AMD’s Zen 5 is a missed opportunity in messaging appeared first on SemiAccurate.
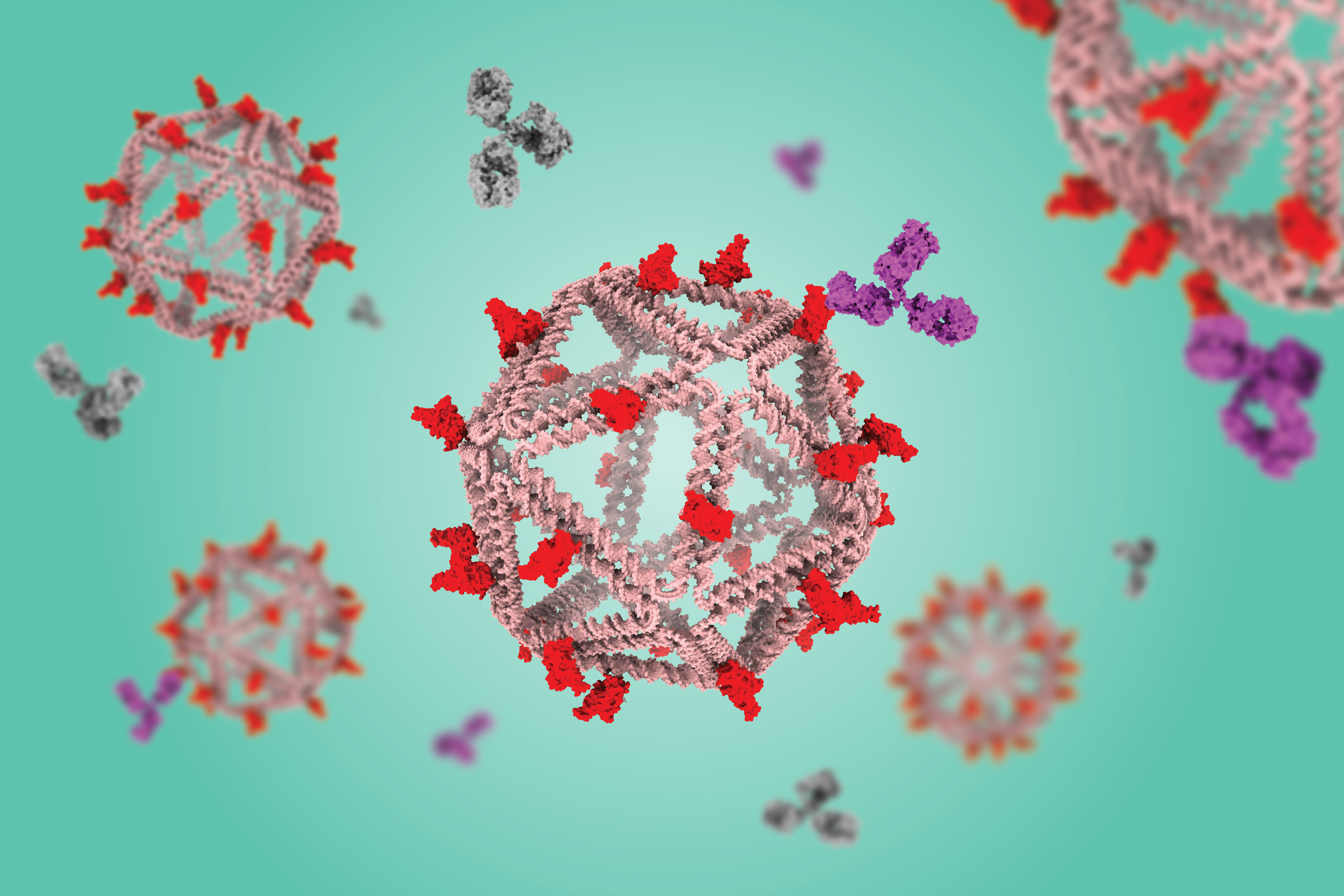
Using a virus-like delivery particle made from DNA, researchers from MIT and the Ragon Institute of MGH, MIT, and Harvard have created a vaccine that can induce a strong antibody response against SARS-CoV-2.
The vaccine, which has been tested in mice, consists of a DNA scaffold that carries many copies of a viral antigen. This type of vaccine, known as a particulate vaccine, mimics the structure of a virus. Most previous work on particulate vaccines has relied on protein scaffolds, but the proteins used in those vaccines tend to generate an unnecessary immune response that can distract the immune system from the target.
In the mouse study, the researchers found that the DNA scaffold does not induce an immune response, allowing the immune system to focus its antibody response on the target antigen.
“DNA, we found in this work, does not elicit antibodies that may distract away from the protein of interest,” says Mark Bathe, an MIT professor of biological engineering. “What you can imagine is that your B cells and immune system are being fully trained by that target antigen, and that’s what you want — for your immune system to be laser-focused on the antigen of interest.”
This approach, which strongly stimulates B cells (the cells that produce antibodies), could make it easier to develop vaccines against viruses that have been difficult to target, including HIV and influenza, as well as SARS-CoV-2, the researchers say. Unlike T cells, which are stimulated by other types of vaccines, these B cells can persist for decades, offering long-term protection.
“We’re interested in exploring whether we can teach the immune system to deliver higher levels of immunity against pathogens that resist conventional vaccine approaches, like flu, HIV, and SARS-CoV-2,” says Daniel Lingwood, an associate professor at Harvard Medical School and a principal investigator at the Ragon Institute. “This idea of decoupling the response against the target antigen from the platform itself is a potentially powerful immunological trick that one can now bring to bear to help those immunological targeting decisions move in a direction that is more focused.”
Bathe, Lingwood, and Aaron Schmidt, an associate professor at Harvard Medical School and principal investigator at the Ragon Institute, are the senior authors of the paper, which appears today in Nature Communications. The paper’s lead authors are Eike-Christian Wamhoff, a former MIT postdoc; Larance Ronsard, a Ragon Institute postdoc; Jared Feldman, a former Harvard University graduate student; Grant Knappe, an MIT graduate student; and Blake Hauser, a former Harvard graduate student.
Mimicking viruses
Particulate vaccines usually consist of a protein nanoparticle, similar in structure to a virus, that can carry many copies of a viral antigen. This high density of antigens can lead to a stronger immune response than traditional vaccines because the body sees it as similar to an actual virus. Particulate vaccines have been developed for a handful of pathogens, including hepatitis B and human papillomavirus, and a particulate vaccine for SARS-CoV-2 has been approved for use in South Korea.
These vaccines are especially good at activating B cells, which produce antibodies specific to the vaccine antigen.
“Particulate vaccines are of great interest for many in immunology because they give you robust humoral immunity, which is antibody-based immunity, which is differentiated from the T-cell-based immunity that the mRNA vaccines seem to elicit more strongly,” Bathe says.
A potential drawback to this kind of vaccine, however, is that the proteins used for the scaffold often stimulate the body to produce antibodies targeting the scaffold. This can distract the immune system and prevent it from launching as robust a response as one would like, Bathe says.
“To neutralize the SARS-CoV-2 virus, you want to have a vaccine that generates antibodies toward the receptor binding domain portion of the virus’ spike protein,” he says. “When you display that on a protein-based particle, what happens is your immune system recognizes not only that receptor binding domain protein, but all the other proteins that are irrelevant to the immune response you’re trying to elicit.”
Another potential drawback is that if the same person receives more than one vaccine carried by the same protein scaffold, for example, SARS-CoV-2 and then influenza, their immune system would likely respond right away to the protein scaffold, having already been primed to react to it. This could weaken the immune response to the antigen carried by the second vaccine.
“If you want to apply that protein-based particle to immunize against a different virus like influenza, then your immune system can be addicted to the underlying protein scaffold that it’s already seen and developed an immune response toward,” Bathe says. “That can hypothetically diminish the quality of your antibody response for the actual antigen of interest.”
As an alternative, Bathe’s lab has been developing scaffolds made using DNA origami, a method that offers precise control over the structure of synthetic DNA and allows researchers to attach a variety of molecules, such as viral antigens, at specific locations.
In a 2020 study, Bathe and Darrell Irvine, an MIT professor of biological engineering and of materials science and engineering, showed that a DNA scaffold carrying 30 copies of an HIV antigen could generate a strong antibody response in B cells grown in the lab. This type of structure is optimal for activating B cells because it closely mimics the structure of nano-sized viruses, which display many copies of viral proteins in their surfaces.
“This approach builds off of a fundamental principle in B-cell antigen recognition, which is that if you have an arrayed display of the antigen, that promotes B-cell responses and gives better quantity and quality of antibody output,” Lingwood says.
“Immunologically silent”
In the new study, the researchers swapped in an antigen consisting of the receptor binding protein of the spike protein from the original strain of SARS-CoV-2. When they gave the vaccine to mice, they found that the mice generated high levels of antibodies to the spike protein but did not generate any to the DNA scaffold.
In contrast, a vaccine based on a scaffold protein called ferritin, coated with SARS-CoV-2 antigens, generated many antibodies against ferritin as well as SARS-CoV-2.
“The DNA nanoparticle itself is immunogenically silent,” Lingwood says. “If you use a protein-based platform, you get equally high titer antibody responses to the platform and to the antigen of interest, and that can complicate repeated usage of that platform because you’ll develop high affinity immune memory against it.”
Reducing these off-target effects could also help scientists reach the goal of developing a vaccine that would induce broadly neutralizing antibodies to any variant of SARS-CoV-2, or even to all sarbecoviruses, the subgenus of virus that includes SARS-CoV-2 as well as the viruses that cause SARS and MERS.
To that end, the researchers are now exploring whether a DNA scaffold with many different viral antigens attached could induce broadly neutralizing antibodies against SARS-CoV-2 and related viruses.
The research was primarily funded by the National Institutes of Health, the National Science Foundation, and the Fast Grants program.
© Credit: The Bathe Lab
Using a virus-like delivery particle made from DNA, researchers from MIT and the Ragon Institute of MGH, MIT, and Harvard have created a vaccine that can induce a strong antibody response against SARS-CoV-2.
The vaccine, which has been tested in mice, consists of a DNA scaffold that carries many copies of a viral antigen. This type of vaccine, known as a particulate vaccine, mimics the structure of a virus. Most previous work on particulate vaccines has relied on protein scaffolds, but the proteins used in those vaccines tend to generate an unnecessary immune response that can distract the immune system from the target.
In the mouse study, the researchers found that the DNA scaffold does not induce an immune response, allowing the immune system to focus its antibody response on the target antigen.
“DNA, we found in this work, does not elicit antibodies that may distract away from the protein of interest,” says Mark Bathe, an MIT professor of biological engineering. “What you can imagine is that your B cells and immune system are being fully trained by that target antigen, and that’s what you want — for your immune system to be laser-focused on the antigen of interest.”
This approach, which strongly stimulates B cells (the cells that produce antibodies), could make it easier to develop vaccines against viruses that have been difficult to target, including HIV and influenza, as well as SARS-CoV-2, the researchers say. Unlike T cells, which are stimulated by other types of vaccines, these B cells can persist for decades, offering long-term protection.
“We’re interested in exploring whether we can teach the immune system to deliver higher levels of immunity against pathogens that resist conventional vaccine approaches, like flu, HIV, and SARS-CoV-2,” says Daniel Lingwood, an associate professor at Harvard Medical School and a principal investigator at the Ragon Institute. “This idea of decoupling the response against the target antigen from the platform itself is a potentially powerful immunological trick that one can now bring to bear to help those immunological targeting decisions move in a direction that is more focused.”
Bathe, Lingwood, and Aaron Schmidt, an associate professor at Harvard Medical School and principal investigator at the Ragon Institute, are the senior authors of the paper, which appears today in Nature Communications. The paper’s lead authors are Eike-Christian Wamhoff, a former MIT postdoc; Larance Ronsard, a Ragon Institute postdoc; Jared Feldman, a former Harvard University graduate student; Grant Knappe, an MIT graduate student; and Blake Hauser, a former Harvard graduate student.
Mimicking viruses
Particulate vaccines usually consist of a protein nanoparticle, similar in structure to a virus, that can carry many copies of a viral antigen. This high density of antigens can lead to a stronger immune response than traditional vaccines because the body sees it as similar to an actual virus. Particulate vaccines have been developed for a handful of pathogens, including hepatitis B and human papillomavirus, and a particulate vaccine for SARS-CoV-2 has been approved for use in South Korea.
These vaccines are especially good at activating B cells, which produce antibodies specific to the vaccine antigen.
“Particulate vaccines are of great interest for many in immunology because they give you robust humoral immunity, which is antibody-based immunity, which is differentiated from the T-cell-based immunity that the mRNA vaccines seem to elicit more strongly,” Bathe says.
A potential drawback to this kind of vaccine, however, is that the proteins used for the scaffold often stimulate the body to produce antibodies targeting the scaffold. This can distract the immune system and prevent it from launching as robust a response as one would like, Bathe says.
“To neutralize the SARS-CoV-2 virus, you want to have a vaccine that generates antibodies toward the receptor binding domain portion of the virus’ spike protein,” he says. “When you display that on a protein-based particle, what happens is your immune system recognizes not only that receptor binding domain protein, but all the other proteins that are irrelevant to the immune response you’re trying to elicit.”
Another potential drawback is that if the same person receives more than one vaccine carried by the same protein scaffold, for example, SARS-CoV-2 and then influenza, their immune system would likely respond right away to the protein scaffold, having already been primed to react to it. This could weaken the immune response to the antigen carried by the second vaccine.
“If you want to apply that protein-based particle to immunize against a different virus like influenza, then your immune system can be addicted to the underlying protein scaffold that it’s already seen and developed an immune response toward,” Bathe says. “That can hypothetically diminish the quality of your antibody response for the actual antigen of interest.”
As an alternative, Bathe’s lab has been developing scaffolds made using DNA origami, a method that offers precise control over the structure of synthetic DNA and allows researchers to attach a variety of molecules, such as viral antigens, at specific locations.
In a 2020 study, Bathe and Darrell Irvine, an MIT professor of biological engineering and of materials science and engineering, showed that a DNA scaffold carrying 30 copies of an HIV antigen could generate a strong antibody response in B cells grown in the lab. This type of structure is optimal for activating B cells because it closely mimics the structure of nano-sized viruses, which display many copies of viral proteins in their surfaces.
“This approach builds off of a fundamental principle in B-cell antigen recognition, which is that if you have an arrayed display of the antigen, that promotes B-cell responses and gives better quantity and quality of antibody output,” Lingwood says.
“Immunologically silent”
In the new study, the researchers swapped in an antigen consisting of the receptor binding protein of the spike protein from the original strain of SARS-CoV-2. When they gave the vaccine to mice, they found that the mice generated high levels of antibodies to the spike protein but did not generate any to the DNA scaffold.
In contrast, a vaccine based on a scaffold protein called ferritin, coated with SARS-CoV-2 antigens, generated many antibodies against ferritin as well as SARS-CoV-2.
“The DNA nanoparticle itself is immunogenically silent,” Lingwood says. “If you use a protein-based platform, you get equally high titer antibody responses to the platform and to the antigen of interest, and that can complicate repeated usage of that platform because you’ll develop high affinity immune memory against it.”
Reducing these off-target effects could also help scientists reach the goal of developing a vaccine that would induce broadly neutralizing antibodies to any variant of SARS-CoV-2, or even to all sarbecoviruses, the subgenus of virus that includes SARS-CoV-2 as well as the viruses that cause SARS and MERS.
To that end, the researchers are now exploring whether a DNA scaffold with many different viral antigens attached could induce broadly neutralizing antibodies against SARS-CoV-2 and related viruses.
The research was primarily funded by the National Institutes of Health, the National Science Foundation, and the Fast Grants program.

© Credit: The Bathe Lab
The state of New Jersey has been sued twice over its infant DNA program. Like the rest of the nation, New Jersey hospitals collect a blood sample from newborns to test them for 60 different health disorders. That part is normal.
But New Jersey is different. Rather than discard the samples after the testing is complete, it holds onto them. For twenty-three years. That’s unusual. And it’s a fair bet that almost 100% of New Jersey parents are unaware of this fact.
There’s a reason parents don’t know this and it has nothing to do with parents just not paying attention when this test is performed. According to the lawsuits, New Jersey healthcare professionals do what they can to portray the testing as mandatory, even though it isn’t. They also take care to keep parents uninformed, never once informing them that they are free to opt out of the testing for religious reasons.
The state, however, is fine with this. The biggest beneficiary of this program is state law enforcement, which can freely obtain these DNA samples without having to go through the trouble of obtaining a warrant. Warrants are needed to obtain DNA samples from criminal suspects, but there’s nothing stopping cops from searching the DNA database for younger relatives of the suspect whose DNA might still be in the possession of the state’s Health Department.
That’s why the state is facing multiple lawsuits, making it an anomaly in this group of 50 states we Americans call home. And that’s likely why the state’s health officials are trying to healthwash this by crafting a new narrative for this uniquely New Jersey handling of infant blood tests. Here’s Elizabeth Nolan Brown with a summary of the rebranding for Reason.
Mandatory genomic sequencing of all newborns—it sounds like something out of a dystopian sci-fi story. But it could become a reality in New Jersey, where health officials are considering adding this analysis to the state’s mandatory newborn testing regime.
Genomic sequencing can determine a person’s “entire genetic makeup,” the National Cancer Institute website explains. Using genomic sequencing, doctors can diagnose diseases and abnormalities, reveal sensitivities to environmental stimulants, and assess a person’s risk of developing conditions such as Alzheimer’s disease.
Ernest Post, chairman of the New Jersey Newborn Screening Advisory Review Committee (NSARC), discussed newborn genomic sequencing at an NSARC meeting in May. An NSARC subcommittee has been convened to explore the issue and is expected to issue recommendations later this year. It’s considering questions such as whether sequencing would be optional or mandatory, the New Jersey Monitor reported.
The state wants to take what’s already problematic and make it a privacy nightmare. But, you know, for the children. The framing encourages people to think this is about early detection and preemptive responses to expected long-term health problems.
And that’s not to stay it won’t have the stated effect. The problem is the state hasn’t been honest about its newborn DNA collection in the past and health care providers (whether ignorant of the facts or instructed to maximize consent) haven’t been exactly trustworthy either.
Now, the state wants to expand what it can do with these blood samples despite not having done anything to correct what’s wrong with the program as it exists already. This just opens up additional avenues of abuse for the government — something it shouldn’t even be considering while it’s still facing two lawsuits related to the existing DNA harvesting program.
The ACLU is obviously opposed to this expansion. The statement it gave to the New Jersey Monitor makes it clear what’s at stake, and what needs to happen before the state moves forward with gene sequencing of newborn blood samples.
If New Jersey adopts genomic sequencing, policymakers must create “a real privacy-protective infrastructure to make sure that genomic data isn’t abused,” said Dillon Reisman, an ACLU-NJ staff attorney.
“What we’re talking about is information from kids that could allow the state and other actors to use that data to monitor and surveil them and their families for the rest of their lives,” Reisman said. “If the goal is the health of children, it does not serve the health of children to have a wild west of genomic data just sitting out there for anyone to abuse.”
Maybe that will happen before this program goes into effect. But it seems unlikely. Given the history of the existing program, the most probable outcome is a handful of alterations as the result of court orders in the lawsuits that are sure to greet the rollout of this program. The state seems super-interested in getting out ahead of health problems. But it seemingly couldn’t care less about heading off the inherent privacy problems the new program would create.
Using a virus-like delivery particle made from DNA, researchers from MIT and the Ragon Institute of MGH, MIT, and Harvard have created a vaccine that can induce a strong antibody response against SARS-CoV-2.
The vaccine, which has been tested in mice, consists of a DNA scaffold that carries many copies of a viral antigen. This type of vaccine, known as a particulate vaccine, mimics the structure of a virus. Most previous work on particulate vaccines has relied on protein scaffolds, but the proteins used in those vaccines tend to generate an unnecessary immune response that can distract the immune system from the target.
In the mouse study, the researchers found that the DNA scaffold does not induce an immune response, allowing the immune system to focus its antibody response on the target antigen.
“DNA, we found in this work, does not elicit antibodies that may distract away from the protein of interest,” says Mark Bathe, an MIT professor of biological engineering. “What you can imagine is that your B cells and immune system are being fully trained by that target antigen, and that’s what you want — for your immune system to be laser-focused on the antigen of interest.”
This approach, which strongly stimulates B cells (the cells that produce antibodies), could make it easier to develop vaccines against viruses that have been difficult to target, including HIV and influenza, as well as SARS-CoV-2, the researchers say. Unlike T cells, which are stimulated by other types of vaccines, these B cells can persist for decades, offering long-term protection.
“We’re interested in exploring whether we can teach the immune system to deliver higher levels of immunity against pathogens that resist conventional vaccine approaches, like flu, HIV, and SARS-CoV-2,” says Daniel Lingwood, an associate professor at Harvard Medical School and a principal investigator at the Ragon Institute. “This idea of decoupling the response against the target antigen from the platform itself is a potentially powerful immunological trick that one can now bring to bear to help those immunological targeting decisions move in a direction that is more focused.”
Bathe, Lingwood, and Aaron Schmidt, an associate professor at Harvard Medical School and principal investigator at the Ragon Institute, are the senior authors of the paper, which appears today in Nature Communications. The paper’s lead authors are Eike-Christian Wamhoff, a former MIT postdoc; Larance Ronsard, a Ragon Institute postdoc; Jared Feldman, a former Harvard University graduate student; Grant Knappe, an MIT graduate student; and Blake Hauser, a former Harvard graduate student.
Mimicking viruses
Particulate vaccines usually consist of a protein nanoparticle, similar in structure to a virus, that can carry many copies of a viral antigen. This high density of antigens can lead to a stronger immune response than traditional vaccines because the body sees it as similar to an actual virus. Particulate vaccines have been developed for a handful of pathogens, including hepatitis B and human papillomavirus, and a particulate vaccine for SARS-CoV-2 has been approved for use in South Korea.
These vaccines are especially good at activating B cells, which produce antibodies specific to the vaccine antigen.
“Particulate vaccines are of great interest for many in immunology because they give you robust humoral immunity, which is antibody-based immunity, which is differentiated from the T-cell-based immunity that the mRNA vaccines seem to elicit more strongly,” Bathe says.
A potential drawback to this kind of vaccine, however, is that the proteins used for the scaffold often stimulate the body to produce antibodies targeting the scaffold. This can distract the immune system and prevent it from launching as robust a response as one would like, Bathe says.
“To neutralize the SARS-CoV-2 virus, you want to have a vaccine that generates antibodies toward the receptor binding domain portion of the virus’ spike protein,” he says. “When you display that on a protein-based particle, what happens is your immune system recognizes not only that receptor binding domain protein, but all the other proteins that are irrelevant to the immune response you’re trying to elicit.”
Another potential drawback is that if the same person receives more than one vaccine carried by the same protein scaffold, for example, SARS-CoV-2 and then influenza, their immune system would likely respond right away to the protein scaffold, having already been primed to react to it. This could weaken the immune response to the antigen carried by the second vaccine.
“If you want to apply that protein-based particle to immunize against a different virus like influenza, then your immune system can be addicted to the underlying protein scaffold that it’s already seen and developed an immune response toward,” Bathe says. “That can hypothetically diminish the quality of your antibody response for the actual antigen of interest.”
As an alternative, Bathe’s lab has been developing scaffolds made using DNA origami, a method that offers precise control over the structure of synthetic DNA and allows researchers to attach a variety of molecules, such as viral antigens, at specific locations.
In a 2020 study, Bathe and Darrell Irvine, an MIT professor of biological engineering and of materials science and engineering, showed that a DNA scaffold carrying 30 copies of an HIV antigen could generate a strong antibody response in B cells grown in the lab. This type of structure is optimal for activating B cells because it closely mimics the structure of nano-sized viruses, which display many copies of viral proteins in their surfaces.
“This approach builds off of a fundamental principle in B-cell antigen recognition, which is that if you have an arrayed display of the antigen, that promotes B-cell responses and gives better quantity and quality of antibody output,” Lingwood says.
“Immunologically silent”
In the new study, the researchers swapped in an antigen consisting of the receptor binding protein of the spike protein from the original strain of SARS-CoV-2. When they gave the vaccine to mice, they found that the mice generated high levels of antibodies to the spike protein but did not generate any to the DNA scaffold.
In contrast, a vaccine based on a scaffold protein called ferritin, coated with SARS-CoV-2 antigens, generated many antibodies against ferritin as well as SARS-CoV-2.
“The DNA nanoparticle itself is immunogenically silent,” Lingwood says. “If you use a protein-based platform, you get equally high titer antibody responses to the platform and to the antigen of interest, and that can complicate repeated usage of that platform because you’ll develop high affinity immune memory against it.”
Reducing these off-target effects could also help scientists reach the goal of developing a vaccine that would induce broadly neutralizing antibodies to any variant of SARS-CoV-2, or even to all sarbecoviruses, the subgenus of virus that includes SARS-CoV-2 as well as the viruses that cause SARS and MERS.
To that end, the researchers are now exploring whether a DNA scaffold with many different viral antigens attached could induce broadly neutralizing antibodies against SARS-CoV-2 and related viruses.
The research was primarily funded by the National Institutes of Health, the National Science Foundation, and the Fast Grants program.

© Credit: The Bathe Lab
Tumors constantly shed DNA from dying cells, which briefly circulates in the patient’s bloodstream before it is quickly broken down. Many companies have created blood tests that can pick out this tumor DNA, potentially helping doctors diagnose or monitor cancer or choose a treatment.
The amount of tumor DNA circulating at any given time, however, is extremely small, so it has been challenging to develop tests sensitive enough to pick up that tiny signal. A team of researchers from MIT and the Broad Institute of MIT and Harvard has now come up with a way to significantly boost that signal, by temporarily slowing the clearance of tumor DNA circulating in the bloodstream.
The researchers developed two different types of injectable molecules that they call “priming agents,” which can transiently interfere with the body’s ability to remove circulating tumor DNA from the bloodstream. In a study of mice, they showed that these agents could boost DNA levels enough that the percentage of detectable early-stage lung metastases leapt from less than 10 percent to above 75 percent.
This approach could enable not only earlier diagnosis of cancer, but also more sensitive detection of tumor mutations that could be used to guide treatment. It could also help improve detection of cancer recurrence.
“You can give one of these agents an hour before the blood draw, and it makes things visible that previously wouldn’t have been. The implication is that we should be able to give everybody who’s doing liquid biopsies, for any purpose, more molecules to work with,” says Sangeeta Bhatia, the John and Dorothy Wilson Professor of Health Sciences and Technology and of Electrical Engineering and Computer Science at MIT, and a member of MIT’s Koch Institute for Integrative Cancer Research and the Institute for Medical Engineering and Science.
Bhatia is one of the senior authors of the new study, along with J. Christopher Love, the Raymond A. and Helen E. St. Laurent Professor of Chemical Engineering at MIT and a member of the Koch Institute and the Ragon Institute of MGH, MIT, and Harvard and Viktor Adalsteinsson, director of the Gerstner Center for Cancer Diagnostics at the Broad Institute.
Carmen Martin-Alonso PhD ’23, MIT and Broad Institute postdoc Shervin Tabrizi, and Broad Institute scientist Kan Xiong are the lead authors of the paper, which appears today in Science.
Better biopsies
Liquid biopsies, which enable detection of small quantities of DNA in blood samples, are now used in many cancer patients to identify mutations that could help guide treatment. With greater sensitivity, however, these tests could become useful for far more patients. Most efforts to improve the sensitivity of liquid biopsies have focused on developing new sequencing technologies to use after the blood is drawn.
While brainstorming ways to make liquid biopsies more informative, Bhatia, Love, Adalsteinsson, and their trainees came up with the idea of trying to increase the amount of DNA in a patient’s bloodstream before the sample is taken.
“A tumor is always creating new cell-free DNA, and that’s the signal that we’re attempting to detect in the blood draw. Existing liquid biopsy technologies, however, are limited by the amount of material you collect in the tube of blood,” Love says. “Where this work intercedes is thinking about how to inject something beforehand that would help boost or enhance the amount of signal that is available to collect in the same small sample.”
The body uses two primary strategies to remove circulating DNA from the bloodstream. Enzymes called DNases circulate in the blood and break down DNA that they encounter, while immune cells known as macrophages take up cell-free DNA as blood is filtered through the liver.
The researchers decided to target each of these processes separately. To prevent DNases from breaking down DNA, they designed a monoclonal antibody that binds to circulating DNA and protects it from the enzymes.
“Antibodies are well-established biopharmaceutical modalities, and they’re safe in a number of different disease contexts, including cancer and autoimmune treatments,” Love says. “The idea was, could we use this kind of antibody to help shield the DNA temporarily from degradation by the nucleases that are in circulation? And by doing so, we shift the balance to where the tumor is generating DNA slightly faster than is being degraded, increasing the concentration in a blood draw.”
The other priming agent they developed is a nanoparticle designed to block macrophages from taking up cell-free DNA. These cells have a well-known tendency to eat up synthetic nanoparticles.
“DNA is a biological nanoparticle, and it made sense that immune cells in the liver were probably taking this up just like they do synthetic nanoparticles. And if that were the case, which it turned out to be, then we could use a safe dummy nanoparticle to distract those immune cells and leave the circulating DNA alone so that it could be at a higher concentration,” Bhatia says.
Earlier tumor detection
The researchers tested their priming agents in mice that received transplants of cancer cells that tend to form tumors in the lungs. Two weeks after the cells were transplanted, the researchers showed that these priming agents could boost the amount of circulating tumor DNA recovered in a blood sample by up to 60-fold.
Once the blood sample is taken, it can be run through the same kinds of sequencing tests now used on liquid biopsy samples. These tests can pick out tumor DNA, including specific sequences used to determine the type of tumor and potentially what kinds of treatments would work best.
Early detection of cancer is another promising application for these priming agents. The researchers found that when mice were given the nanoparticle priming agent before blood was drawn, it allowed them to detect circulating tumor DNA in blood of 75 percent of the mice with low cancer burden, while none were detectable without this boost.
“One of the greatest hurdles for cancer liquid biopsy testing has been the scarcity of circulating tumor DNA in a blood sample,” Adalsteinsson says. “It’s thus been encouraging to see the magnitude of the effect we’ve been able to achieve so far and to envision what impact this could have for patients.”
After either of the priming agents are injected, it takes an hour or two for the DNA levels to increase in the bloodstream, and then they return to normal within about 24 hours.
“The ability to get peak activity of these agents within a couple of hours, followed by their rapid clearance, means that someone could go into a doctor’s office, receive an agent like this, and then give their blood for the test itself, all within one visit,” Love says. “This feature bodes well for the potential to translate this concept into clinical use.”
The researchers have launched a company called Amplifyer Bio that plans to further develop the technology, in hopes of advancing to clinical trials.
“A tube of blood is a much more accessible diagnostic than colonoscopy screening or even mammography,” Bhatia says. “Ultimately, if these tools really are predictive, then we should be able to get many more patients into the system who could benefit from cancer interception or better therapy.”
The research was funded by the Koch Institute Support (core) Grant from the National Cancer Institute, the Marble Center for Cancer Nanomedicine, the Gerstner Family Foundation, the Ludwig Center at MIT, the Koch Institute Frontier Research Program via the Casey and Family Foundation, and the Bridge Project, a partnership between the Koch Institute and the Dana-Farber/Harvard Cancer Center.

© Image: MIT News; iStock
Using a virus-like delivery particle made from DNA, researchers from MIT and the Ragon Institute of MGH, MIT, and Harvard have created a vaccine that can induce a strong antibody response against SARS-CoV-2.
The vaccine, which has been tested in mice, consists of a DNA scaffold that carries many copies of a viral antigen. This type of vaccine, known as a particulate vaccine, mimics the structure of a virus. Most previous work on particulate vaccines has relied on protein scaffolds, but the proteins used in those vaccines tend to generate an unnecessary immune response that can distract the immune system from the target.
In the mouse study, the researchers found that the DNA scaffold does not induce an immune response, allowing the immune system to focus its antibody response on the target antigen.
“DNA, we found in this work, does not elicit antibodies that may distract away from the protein of interest,” says Mark Bathe, an MIT professor of biological engineering. “What you can imagine is that your B cells and immune system are being fully trained by that target antigen, and that’s what you want — for your immune system to be laser-focused on the antigen of interest.”
This approach, which strongly stimulates B cells (the cells that produce antibodies), could make it easier to develop vaccines against viruses that have been difficult to target, including HIV and influenza, as well as SARS-CoV-2, the researchers say. Unlike T cells, which are stimulated by other types of vaccines, these B cells can persist for decades, offering long-term protection.
“We’re interested in exploring whether we can teach the immune system to deliver higher levels of immunity against pathogens that resist conventional vaccine approaches, like flu, HIV, and SARS-CoV-2,” says Daniel Lingwood, an associate professor at Harvard Medical School and a principal investigator at the Ragon Institute. “This idea of decoupling the response against the target antigen from the platform itself is a potentially powerful immunological trick that one can now bring to bear to help those immunological targeting decisions move in a direction that is more focused.”
Bathe, Lingwood, and Aaron Schmidt, an associate professor at Harvard Medical School and principal investigator at the Ragon Institute, are the senior authors of the paper, which appears today in Nature Communications. The paper’s lead authors are Eike-Christian Wamhoff, a former MIT postdoc; Larance Ronsard, a Ragon Institute postdoc; Jared Feldman, a former Harvard University graduate student; Grant Knappe, an MIT graduate student; and Blake Hauser, a former Harvard graduate student.
Mimicking viruses
Particulate vaccines usually consist of a protein nanoparticle, similar in structure to a virus, that can carry many copies of a viral antigen. This high density of antigens can lead to a stronger immune response than traditional vaccines because the body sees it as similar to an actual virus. Particulate vaccines have been developed for a handful of pathogens, including hepatitis B and human papillomavirus, and a particulate vaccine for SARS-CoV-2 has been approved for use in South Korea.
These vaccines are especially good at activating B cells, which produce antibodies specific to the vaccine antigen.
“Particulate vaccines are of great interest for many in immunology because they give you robust humoral immunity, which is antibody-based immunity, which is differentiated from the T-cell-based immunity that the mRNA vaccines seem to elicit more strongly,” Bathe says.
A potential drawback to this kind of vaccine, however, is that the proteins used for the scaffold often stimulate the body to produce antibodies targeting the scaffold. This can distract the immune system and prevent it from launching as robust a response as one would like, Bathe says.
“To neutralize the SARS-CoV-2 virus, you want to have a vaccine that generates antibodies toward the receptor binding domain portion of the virus’ spike protein,” he says. “When you display that on a protein-based particle, what happens is your immune system recognizes not only that receptor binding domain protein, but all the other proteins that are irrelevant to the immune response you’re trying to elicit.”
Another potential drawback is that if the same person receives more than one vaccine carried by the same protein scaffold, for example, SARS-CoV-2 and then influenza, their immune system would likely respond right away to the protein scaffold, having already been primed to react to it. This could weaken the immune response to the antigen carried by the second vaccine.
“If you want to apply that protein-based particle to immunize against a different virus like influenza, then your immune system can be addicted to the underlying protein scaffold that it’s already seen and developed an immune response toward,” Bathe says. “That can hypothetically diminish the quality of your antibody response for the actual antigen of interest.”
As an alternative, Bathe’s lab has been developing scaffolds made using DNA origami, a method that offers precise control over the structure of synthetic DNA and allows researchers to attach a variety of molecules, such as viral antigens, at specific locations.
In a 2020 study, Bathe and Darrell Irvine, an MIT professor of biological engineering and of materials science and engineering, showed that a DNA scaffold carrying 30 copies of an HIV antigen could generate a strong antibody response in B cells grown in the lab. This type of structure is optimal for activating B cells because it closely mimics the structure of nano-sized viruses, which display many copies of viral proteins in their surfaces.
“This approach builds off of a fundamental principle in B-cell antigen recognition, which is that if you have an arrayed display of the antigen, that promotes B-cell responses and gives better quantity and quality of antibody output,” Lingwood says.
“Immunologically silent”
In the new study, the researchers swapped in an antigen consisting of the receptor binding protein of the spike protein from the original strain of SARS-CoV-2. When they gave the vaccine to mice, they found that the mice generated high levels of antibodies to the spike protein but did not generate any to the DNA scaffold.
In contrast, a vaccine based on a scaffold protein called ferritin, coated with SARS-CoV-2 antigens, generated many antibodies against ferritin as well as SARS-CoV-2.
“The DNA nanoparticle itself is immunogenically silent,” Lingwood says. “If you use a protein-based platform, you get equally high titer antibody responses to the platform and to the antigen of interest, and that can complicate repeated usage of that platform because you’ll develop high affinity immune memory against it.”
Reducing these off-target effects could also help scientists reach the goal of developing a vaccine that would induce broadly neutralizing antibodies to any variant of SARS-CoV-2, or even to all sarbecoviruses, the subgenus of virus that includes SARS-CoV-2 as well as the viruses that cause SARS and MERS.
To that end, the researchers are now exploring whether a DNA scaffold with many different viral antigens attached could induce broadly neutralizing antibodies against SARS-CoV-2 and related viruses.
The research was primarily funded by the National Institutes of Health, the National Science Foundation, and the Fast Grants program.

© Credit: The Bathe Lab
Tumors constantly shed DNA from dying cells, which briefly circulates in the patient’s bloodstream before it is quickly broken down. Many companies have created blood tests that can pick out this tumor DNA, potentially helping doctors diagnose or monitor cancer or choose a treatment.
The amount of tumor DNA circulating at any given time, however, is extremely small, so it has been challenging to develop tests sensitive enough to pick up that tiny signal. A team of researchers from MIT and the Broad Institute of MIT and Harvard has now come up with a way to significantly boost that signal, by temporarily slowing the clearance of tumor DNA circulating in the bloodstream.
The researchers developed two different types of injectable molecules that they call “priming agents,” which can transiently interfere with the body’s ability to remove circulating tumor DNA from the bloodstream. In a study of mice, they showed that these agents could boost DNA levels enough that the percentage of detectable early-stage lung metastases leapt from less than 10 percent to above 75 percent.
This approach could enable not only earlier diagnosis of cancer, but also more sensitive detection of tumor mutations that could be used to guide treatment. It could also help improve detection of cancer recurrence.
“You can give one of these agents an hour before the blood draw, and it makes things visible that previously wouldn’t have been. The implication is that we should be able to give everybody who’s doing liquid biopsies, for any purpose, more molecules to work with,” says Sangeeta Bhatia, the John and Dorothy Wilson Professor of Health Sciences and Technology and of Electrical Engineering and Computer Science at MIT, and a member of MIT’s Koch Institute for Integrative Cancer Research and the Institute for Medical Engineering and Science.
Bhatia is one of the senior authors of the new study, along with J. Christopher Love, the Raymond A. and Helen E. St. Laurent Professor of Chemical Engineering at MIT and a member of the Koch Institute and the Ragon Institute of MGH, MIT, and Harvard and Viktor Adalsteinsson, director of the Gerstner Center for Cancer Diagnostics at the Broad Institute.
Carmen Martin-Alonso PhD ’23, MIT and Broad Institute postdoc Shervin Tabrizi, and Broad Institute scientist Kan Xiong are the lead authors of the paper, which appears today in Science.
Better biopsies
Liquid biopsies, which enable detection of small quantities of DNA in blood samples, are now used in many cancer patients to identify mutations that could help guide treatment. With greater sensitivity, however, these tests could become useful for far more patients. Most efforts to improve the sensitivity of liquid biopsies have focused on developing new sequencing technologies to use after the blood is drawn.
While brainstorming ways to make liquid biopsies more informative, Bhatia, Love, Adalsteinsson, and their trainees came up with the idea of trying to increase the amount of DNA in a patient’s bloodstream before the sample is taken.
“A tumor is always creating new cell-free DNA, and that’s the signal that we’re attempting to detect in the blood draw. Existing liquid biopsy technologies, however, are limited by the amount of material you collect in the tube of blood,” Love says. “Where this work intercedes is thinking about how to inject something beforehand that would help boost or enhance the amount of signal that is available to collect in the same small sample.”
The body uses two primary strategies to remove circulating DNA from the bloodstream. Enzymes called DNases circulate in the blood and break down DNA that they encounter, while immune cells known as macrophages take up cell-free DNA as blood is filtered through the liver.
The researchers decided to target each of these processes separately. To prevent DNases from breaking down DNA, they designed a monoclonal antibody that binds to circulating DNA and protects it from the enzymes.
“Antibodies are well-established biopharmaceutical modalities, and they’re safe in a number of different disease contexts, including cancer and autoimmune treatments,” Love says. “The idea was, could we use this kind of antibody to help shield the DNA temporarily from degradation by the nucleases that are in circulation? And by doing so, we shift the balance to where the tumor is generating DNA slightly faster than is being degraded, increasing the concentration in a blood draw.”
The other priming agent they developed is a nanoparticle designed to block macrophages from taking up cell-free DNA. These cells have a well-known tendency to eat up synthetic nanoparticles.
“DNA is a biological nanoparticle, and it made sense that immune cells in the liver were probably taking this up just like they do synthetic nanoparticles. And if that were the case, which it turned out to be, then we could use a safe dummy nanoparticle to distract those immune cells and leave the circulating DNA alone so that it could be at a higher concentration,” Bhatia says.
Earlier tumor detection
The researchers tested their priming agents in mice that received transplants of cancer cells that tend to form tumors in the lungs. Two weeks after the cells were transplanted, the researchers showed that these priming agents could boost the amount of circulating tumor DNA recovered in a blood sample by up to 60-fold.
Once the blood sample is taken, it can be run through the same kinds of sequencing tests now used on liquid biopsy samples. These tests can pick out tumor DNA, including specific sequences used to determine the type of tumor and potentially what kinds of treatments would work best.
Early detection of cancer is another promising application for these priming agents. The researchers found that when mice were given the nanoparticle priming agent before blood was drawn, it allowed them to detect circulating tumor DNA in blood of 75 percent of the mice with low cancer burden, while none were detectable without this boost.
“One of the greatest hurdles for cancer liquid biopsy testing has been the scarcity of circulating tumor DNA in a blood sample,” Adalsteinsson says. “It’s thus been encouraging to see the magnitude of the effect we’ve been able to achieve so far and to envision what impact this could have for patients.”
After either of the priming agents are injected, it takes an hour or two for the DNA levels to increase in the bloodstream, and then they return to normal within about 24 hours.
“The ability to get peak activity of these agents within a couple of hours, followed by their rapid clearance, means that someone could go into a doctor’s office, receive an agent like this, and then give their blood for the test itself, all within one visit,” Love says. “This feature bodes well for the potential to translate this concept into clinical use.”
The researchers have launched a company called Amplifyer Bio that plans to further develop the technology, in hopes of advancing to clinical trials.
“A tube of blood is a much more accessible diagnostic than colonoscopy screening or even mammography,” Bhatia says. “Ultimately, if these tools really are predictive, then we should be able to get many more patients into the system who could benefit from cancer interception or better therapy.”
The research was funded by the Koch Institute Support (core) Grant from the National Cancer Institute, the Marble Center for Cancer Nanomedicine, the Gerstner Family Foundation, the Ludwig Center at MIT, the Koch Institute Frontier Research Program via the Casey and Family Foundation, and the Bridge Project, a partnership between the Koch Institute and the Dana-Farber/Harvard Cancer Center.

© Image: MIT News; iStock
Using a virus-like delivery particle made from DNA, researchers from MIT and the Ragon Institute of MGH, MIT, and Harvard have created a vaccine that can induce a strong antibody response against SARS-CoV-2.
The vaccine, which has been tested in mice, consists of a DNA scaffold that carries many copies of a viral antigen. This type of vaccine, known as a particulate vaccine, mimics the structure of a virus. Most previous work on particulate vaccines has relied on protein scaffolds, but the proteins used in those vaccines tend to generate an unnecessary immune response that can distract the immune system from the target.
In the mouse study, the researchers found that the DNA scaffold does not induce an immune response, allowing the immune system to focus its antibody response on the target antigen.
“DNA, we found in this work, does not elicit antibodies that may distract away from the protein of interest,” says Mark Bathe, an MIT professor of biological engineering. “What you can imagine is that your B cells and immune system are being fully trained by that target antigen, and that’s what you want — for your immune system to be laser-focused on the antigen of interest.”
This approach, which strongly stimulates B cells (the cells that produce antibodies), could make it easier to develop vaccines against viruses that have been difficult to target, including HIV and influenza, as well as SARS-CoV-2, the researchers say. Unlike T cells, which are stimulated by other types of vaccines, these B cells can persist for decades, offering long-term protection.
“We’re interested in exploring whether we can teach the immune system to deliver higher levels of immunity against pathogens that resist conventional vaccine approaches, like flu, HIV, and SARS-CoV-2,” says Daniel Lingwood, an associate professor at Harvard Medical School and a principal investigator at the Ragon Institute. “This idea of decoupling the response against the target antigen from the platform itself is a potentially powerful immunological trick that one can now bring to bear to help those immunological targeting decisions move in a direction that is more focused.”
Bathe, Lingwood, and Aaron Schmidt, an associate professor at Harvard Medical School and principal investigator at the Ragon Institute, are the senior authors of the paper, which appears today in Nature Communications. The paper’s lead authors are Eike-Christian Wamhoff, a former MIT postdoc; Larance Ronsard, a Ragon Institute postdoc; Jared Feldman, a former Harvard University graduate student; Grant Knappe, an MIT graduate student; and Blake Hauser, a former Harvard graduate student.
Mimicking viruses
Particulate vaccines usually consist of a protein nanoparticle, similar in structure to a virus, that can carry many copies of a viral antigen. This high density of antigens can lead to a stronger immune response than traditional vaccines because the body sees it as similar to an actual virus. Particulate vaccines have been developed for a handful of pathogens, including hepatitis B and human papillomavirus, and a particulate vaccine for SARS-CoV-2 has been approved for use in South Korea.
These vaccines are especially good at activating B cells, which produce antibodies specific to the vaccine antigen.
“Particulate vaccines are of great interest for many in immunology because they give you robust humoral immunity, which is antibody-based immunity, which is differentiated from the T-cell-based immunity that the mRNA vaccines seem to elicit more strongly,” Bathe says.
A potential drawback to this kind of vaccine, however, is that the proteins used for the scaffold often stimulate the body to produce antibodies targeting the scaffold. This can distract the immune system and prevent it from launching as robust a response as one would like, Bathe says.
“To neutralize the SARS-CoV-2 virus, you want to have a vaccine that generates antibodies toward the receptor binding domain portion of the virus’ spike protein,” he says. “When you display that on a protein-based particle, what happens is your immune system recognizes not only that receptor binding domain protein, but all the other proteins that are irrelevant to the immune response you’re trying to elicit.”
Another potential drawback is that if the same person receives more than one vaccine carried by the same protein scaffold, for example, SARS-CoV-2 and then influenza, their immune system would likely respond right away to the protein scaffold, having already been primed to react to it. This could weaken the immune response to the antigen carried by the second vaccine.
“If you want to apply that protein-based particle to immunize against a different virus like influenza, then your immune system can be addicted to the underlying protein scaffold that it’s already seen and developed an immune response toward,” Bathe says. “That can hypothetically diminish the quality of your antibody response for the actual antigen of interest.”
As an alternative, Bathe’s lab has been developing scaffolds made using DNA origami, a method that offers precise control over the structure of synthetic DNA and allows researchers to attach a variety of molecules, such as viral antigens, at specific locations.
In a 2020 study, Bathe and Darrell Irvine, an MIT professor of biological engineering and of materials science and engineering, showed that a DNA scaffold carrying 30 copies of an HIV antigen could generate a strong antibody response in B cells grown in the lab. This type of structure is optimal for activating B cells because it closely mimics the structure of nano-sized viruses, which display many copies of viral proteins in their surfaces.
“This approach builds off of a fundamental principle in B-cell antigen recognition, which is that if you have an arrayed display of the antigen, that promotes B-cell responses and gives better quantity and quality of antibody output,” Lingwood says.
“Immunologically silent”
In the new study, the researchers swapped in an antigen consisting of the receptor binding protein of the spike protein from the original strain of SARS-CoV-2. When they gave the vaccine to mice, they found that the mice generated high levels of antibodies to the spike protein but did not generate any to the DNA scaffold.
In contrast, a vaccine based on a scaffold protein called ferritin, coated with SARS-CoV-2 antigens, generated many antibodies against ferritin as well as SARS-CoV-2.
“The DNA nanoparticle itself is immunogenically silent,” Lingwood says. “If you use a protein-based platform, you get equally high titer antibody responses to the platform and to the antigen of interest, and that can complicate repeated usage of that platform because you’ll develop high affinity immune memory against it.”
Reducing these off-target effects could also help scientists reach the goal of developing a vaccine that would induce broadly neutralizing antibodies to any variant of SARS-CoV-2, or even to all sarbecoviruses, the subgenus of virus that includes SARS-CoV-2 as well as the viruses that cause SARS and MERS.
To that end, the researchers are now exploring whether a DNA scaffold with many different viral antigens attached could induce broadly neutralizing antibodies against SARS-CoV-2 and related viruses.
The research was primarily funded by the National Institutes of Health, the National Science Foundation, and the Fast Grants program.

© Credit: The Bathe Lab
Tumors constantly shed DNA from dying cells, which briefly circulates in the patient’s bloodstream before it is quickly broken down. Many companies have created blood tests that can pick out this tumor DNA, potentially helping doctors diagnose or monitor cancer or choose a treatment.
The amount of tumor DNA circulating at any given time, however, is extremely small, so it has been challenging to develop tests sensitive enough to pick up that tiny signal. A team of researchers from MIT and the Broad Institute of MIT and Harvard has now come up with a way to significantly boost that signal, by temporarily slowing the clearance of tumor DNA circulating in the bloodstream.
The researchers developed two different types of injectable molecules that they call “priming agents,” which can transiently interfere with the body’s ability to remove circulating tumor DNA from the bloodstream. In a study of mice, they showed that these agents could boost DNA levels enough that the percentage of detectable early-stage lung metastases leapt from less than 10 percent to above 75 percent.
This approach could enable not only earlier diagnosis of cancer, but also more sensitive detection of tumor mutations that could be used to guide treatment. It could also help improve detection of cancer recurrence.
“You can give one of these agents an hour before the blood draw, and it makes things visible that previously wouldn’t have been. The implication is that we should be able to give everybody who’s doing liquid biopsies, for any purpose, more molecules to work with,” says Sangeeta Bhatia, the John and Dorothy Wilson Professor of Health Sciences and Technology and of Electrical Engineering and Computer Science at MIT, and a member of MIT’s Koch Institute for Integrative Cancer Research and the Institute for Medical Engineering and Science.
Bhatia is one of the senior authors of the new study, along with J. Christopher Love, the Raymond A. and Helen E. St. Laurent Professor of Chemical Engineering at MIT and a member of the Koch Institute and the Ragon Institute of MGH, MIT, and Harvard and Viktor Adalsteinsson, director of the Gerstner Center for Cancer Diagnostics at the Broad Institute.
Carmen Martin-Alonso PhD ’23, MIT and Broad Institute postdoc Shervin Tabrizi, and Broad Institute scientist Kan Xiong are the lead authors of the paper, which appears today in Science.
Better biopsies
Liquid biopsies, which enable detection of small quantities of DNA in blood samples, are now used in many cancer patients to identify mutations that could help guide treatment. With greater sensitivity, however, these tests could become useful for far more patients. Most efforts to improve the sensitivity of liquid biopsies have focused on developing new sequencing technologies to use after the blood is drawn.
While brainstorming ways to make liquid biopsies more informative, Bhatia, Love, Adalsteinsson, and their trainees came up with the idea of trying to increase the amount of DNA in a patient’s bloodstream before the sample is taken.
“A tumor is always creating new cell-free DNA, and that’s the signal that we’re attempting to detect in the blood draw. Existing liquid biopsy technologies, however, are limited by the amount of material you collect in the tube of blood,” Love says. “Where this work intercedes is thinking about how to inject something beforehand that would help boost or enhance the amount of signal that is available to collect in the same small sample.”
The body uses two primary strategies to remove circulating DNA from the bloodstream. Enzymes called DNases circulate in the blood and break down DNA that they encounter, while immune cells known as macrophages take up cell-free DNA as blood is filtered through the liver.
The researchers decided to target each of these processes separately. To prevent DNases from breaking down DNA, they designed a monoclonal antibody that binds to circulating DNA and protects it from the enzymes.
“Antibodies are well-established biopharmaceutical modalities, and they’re safe in a number of different disease contexts, including cancer and autoimmune treatments,” Love says. “The idea was, could we use this kind of antibody to help shield the DNA temporarily from degradation by the nucleases that are in circulation? And by doing so, we shift the balance to where the tumor is generating DNA slightly faster than is being degraded, increasing the concentration in a blood draw.”
The other priming agent they developed is a nanoparticle designed to block macrophages from taking up cell-free DNA. These cells have a well-known tendency to eat up synthetic nanoparticles.
“DNA is a biological nanoparticle, and it made sense that immune cells in the liver were probably taking this up just like they do synthetic nanoparticles. And if that were the case, which it turned out to be, then we could use a safe dummy nanoparticle to distract those immune cells and leave the circulating DNA alone so that it could be at a higher concentration,” Bhatia says.
Earlier tumor detection
The researchers tested their priming agents in mice that received transplants of cancer cells that tend to form tumors in the lungs. Two weeks after the cells were transplanted, the researchers showed that these priming agents could boost the amount of circulating tumor DNA recovered in a blood sample by up to 60-fold.
Once the blood sample is taken, it can be run through the same kinds of sequencing tests now used on liquid biopsy samples. These tests can pick out tumor DNA, including specific sequences used to determine the type of tumor and potentially what kinds of treatments would work best.
Early detection of cancer is another promising application for these priming agents. The researchers found that when mice were given the nanoparticle priming agent before blood was drawn, it allowed them to detect circulating tumor DNA in blood of 75 percent of the mice with low cancer burden, while none were detectable without this boost.
“One of the greatest hurdles for cancer liquid biopsy testing has been the scarcity of circulating tumor DNA in a blood sample,” Adalsteinsson says. “It’s thus been encouraging to see the magnitude of the effect we’ve been able to achieve so far and to envision what impact this could have for patients.”
After either of the priming agents are injected, it takes an hour or two for the DNA levels to increase in the bloodstream, and then they return to normal within about 24 hours.
“The ability to get peak activity of these agents within a couple of hours, followed by their rapid clearance, means that someone could go into a doctor’s office, receive an agent like this, and then give their blood for the test itself, all within one visit,” Love says. “This feature bodes well for the potential to translate this concept into clinical use.”
The researchers have launched a company called Amplifyer Bio that plans to further develop the technology, in hopes of advancing to clinical trials.
“A tube of blood is a much more accessible diagnostic than colonoscopy screening or even mammography,” Bhatia says. “Ultimately, if these tools really are predictive, then we should be able to get many more patients into the system who could benefit from cancer interception or better therapy.”
The research was funded by the Koch Institute Support (core) Grant from the National Cancer Institute, the Marble Center for Cancer Nanomedicine, the Gerstner Family Foundation, the Ludwig Center at MIT, the Koch Institute Frontier Research Program via the Casey and Family Foundation, and the Bridge Project, a partnership between the Koch Institute and the Dana-Farber/Harvard Cancer Center.

© Image: MIT News; iStock

Sonic the Hedgehog 3 is still seven months away, as the third live-action movie starring the blue blur and his angsty rival Shadow will premiere on December 20. And fans are eagerly anticipating the first trailer for the upcoming movie. Though some lucky ones were able to get a first look at a private event in April,…
Using a virus-like delivery particle made from DNA, researchers from MIT and the Ragon Institute of MGH, MIT, and Harvard have created a vaccine that can induce a strong antibody response against SARS-CoV-2.
The vaccine, which has been tested in mice, consists of a DNA scaffold that carries many copies of a viral antigen. This type of vaccine, known as a particulate vaccine, mimics the structure of a virus. Most previous work on particulate vaccines has relied on protein scaffolds, but the proteins used in those vaccines tend to generate an unnecessary immune response that can distract the immune system from the target.
In the mouse study, the researchers found that the DNA scaffold does not induce an immune response, allowing the immune system to focus its antibody response on the target antigen.
“DNA, we found in this work, does not elicit antibodies that may distract away from the protein of interest,” says Mark Bathe, an MIT professor of biological engineering. “What you can imagine is that your B cells and immune system are being fully trained by that target antigen, and that’s what you want — for your immune system to be laser-focused on the antigen of interest.”
This approach, which strongly stimulates B cells (the cells that produce antibodies), could make it easier to develop vaccines against viruses that have been difficult to target, including HIV and influenza, as well as SARS-CoV-2, the researchers say. Unlike T cells, which are stimulated by other types of vaccines, these B cells can persist for decades, offering long-term protection.
“We’re interested in exploring whether we can teach the immune system to deliver higher levels of immunity against pathogens that resist conventional vaccine approaches, like flu, HIV, and SARS-CoV-2,” says Daniel Lingwood, an associate professor at Harvard Medical School and a principal investigator at the Ragon Institute. “This idea of decoupling the response against the target antigen from the platform itself is a potentially powerful immunological trick that one can now bring to bear to help those immunological targeting decisions move in a direction that is more focused.”
Bathe, Lingwood, and Aaron Schmidt, an associate professor at Harvard Medical School and principal investigator at the Ragon Institute, are the senior authors of the paper, which appears today in Nature Communications. The paper’s lead authors are Eike-Christian Wamhoff, a former MIT postdoc; Larance Ronsard, a Ragon Institute postdoc; Jared Feldman, a former Harvard University graduate student; Grant Knappe, an MIT graduate student; and Blake Hauser, a former Harvard graduate student.
Mimicking viruses
Particulate vaccines usually consist of a protein nanoparticle, similar in structure to a virus, that can carry many copies of a viral antigen. This high density of antigens can lead to a stronger immune response than traditional vaccines because the body sees it as similar to an actual virus. Particulate vaccines have been developed for a handful of pathogens, including hepatitis B and human papillomavirus, and a particulate vaccine for SARS-CoV-2 has been approved for use in South Korea.
These vaccines are especially good at activating B cells, which produce antibodies specific to the vaccine antigen.
“Particulate vaccines are of great interest for many in immunology because they give you robust humoral immunity, which is antibody-based immunity, which is differentiated from the T-cell-based immunity that the mRNA vaccines seem to elicit more strongly,” Bathe says.
A potential drawback to this kind of vaccine, however, is that the proteins used for the scaffold often stimulate the body to produce antibodies targeting the scaffold. This can distract the immune system and prevent it from launching as robust a response as one would like, Bathe says.
“To neutralize the SARS-CoV-2 virus, you want to have a vaccine that generates antibodies toward the receptor binding domain portion of the virus’ spike protein,” he says. “When you display that on a protein-based particle, what happens is your immune system recognizes not only that receptor binding domain protein, but all the other proteins that are irrelevant to the immune response you’re trying to elicit.”
Another potential drawback is that if the same person receives more than one vaccine carried by the same protein scaffold, for example, SARS-CoV-2 and then influenza, their immune system would likely respond right away to the protein scaffold, having already been primed to react to it. This could weaken the immune response to the antigen carried by the second vaccine.
“If you want to apply that protein-based particle to immunize against a different virus like influenza, then your immune system can be addicted to the underlying protein scaffold that it’s already seen and developed an immune response toward,” Bathe says. “That can hypothetically diminish the quality of your antibody response for the actual antigen of interest.”
As an alternative, Bathe’s lab has been developing scaffolds made using DNA origami, a method that offers precise control over the structure of synthetic DNA and allows researchers to attach a variety of molecules, such as viral antigens, at specific locations.
In a 2020 study, Bathe and Darrell Irvine, an MIT professor of biological engineering and of materials science and engineering, showed that a DNA scaffold carrying 30 copies of an HIV antigen could generate a strong antibody response in B cells grown in the lab. This type of structure is optimal for activating B cells because it closely mimics the structure of nano-sized viruses, which display many copies of viral proteins in their surfaces.
“This approach builds off of a fundamental principle in B-cell antigen recognition, which is that if you have an arrayed display of the antigen, that promotes B-cell responses and gives better quantity and quality of antibody output,” Lingwood says.
“Immunologically silent”
In the new study, the researchers swapped in an antigen consisting of the receptor binding protein of the spike protein from the original strain of SARS-CoV-2. When they gave the vaccine to mice, they found that the mice generated high levels of antibodies to the spike protein but did not generate any to the DNA scaffold.
In contrast, a vaccine based on a scaffold protein called ferritin, coated with SARS-CoV-2 antigens, generated many antibodies against ferritin as well as SARS-CoV-2.
“The DNA nanoparticle itself is immunogenically silent,” Lingwood says. “If you use a protein-based platform, you get equally high titer antibody responses to the platform and to the antigen of interest, and that can complicate repeated usage of that platform because you’ll develop high affinity immune memory against it.”
Reducing these off-target effects could also help scientists reach the goal of developing a vaccine that would induce broadly neutralizing antibodies to any variant of SARS-CoV-2, or even to all sarbecoviruses, the subgenus of virus that includes SARS-CoV-2 as well as the viruses that cause SARS and MERS.
To that end, the researchers are now exploring whether a DNA scaffold with many different viral antigens attached could induce broadly neutralizing antibodies against SARS-CoV-2 and related viruses.
The research was primarily funded by the National Institutes of Health, the National Science Foundation, and the Fast Grants program.

© Credit: The Bathe Lab
Tumors constantly shed DNA from dying cells, which briefly circulates in the patient’s bloodstream before it is quickly broken down. Many companies have created blood tests that can pick out this tumor DNA, potentially helping doctors diagnose or monitor cancer or choose a treatment.
The amount of tumor DNA circulating at any given time, however, is extremely small, so it has been challenging to develop tests sensitive enough to pick up that tiny signal. A team of researchers from MIT and the Broad Institute of MIT and Harvard has now come up with a way to significantly boost that signal, by temporarily slowing the clearance of tumor DNA circulating in the bloodstream.
The researchers developed two different types of injectable molecules that they call “priming agents,” which can transiently interfere with the body’s ability to remove circulating tumor DNA from the bloodstream. In a study of mice, they showed that these agents could boost DNA levels enough that the percentage of detectable early-stage lung metastases leapt from less than 10 percent to above 75 percent.
This approach could enable not only earlier diagnosis of cancer, but also more sensitive detection of tumor mutations that could be used to guide treatment. It could also help improve detection of cancer recurrence.
“You can give one of these agents an hour before the blood draw, and it makes things visible that previously wouldn’t have been. The implication is that we should be able to give everybody who’s doing liquid biopsies, for any purpose, more molecules to work with,” says Sangeeta Bhatia, the John and Dorothy Wilson Professor of Health Sciences and Technology and of Electrical Engineering and Computer Science at MIT, and a member of MIT’s Koch Institute for Integrative Cancer Research and the Institute for Medical Engineering and Science.
Bhatia is one of the senior authors of the new study, along with J. Christopher Love, the Raymond A. and Helen E. St. Laurent Professor of Chemical Engineering at MIT and a member of the Koch Institute and the Ragon Institute of MGH, MIT, and Harvard and Viktor Adalsteinsson, director of the Gerstner Center for Cancer Diagnostics at the Broad Institute.
Carmen Martin-Alonso PhD ’23, MIT and Broad Institute postdoc Shervin Tabrizi, and Broad Institute scientist Kan Xiong are the lead authors of the paper, which appears today in Science.
Better biopsies
Liquid biopsies, which enable detection of small quantities of DNA in blood samples, are now used in many cancer patients to identify mutations that could help guide treatment. With greater sensitivity, however, these tests could become useful for far more patients. Most efforts to improve the sensitivity of liquid biopsies have focused on developing new sequencing technologies to use after the blood is drawn.
While brainstorming ways to make liquid biopsies more informative, Bhatia, Love, Adalsteinsson, and their trainees came up with the idea of trying to increase the amount of DNA in a patient’s bloodstream before the sample is taken.
“A tumor is always creating new cell-free DNA, and that’s the signal that we’re attempting to detect in the blood draw. Existing liquid biopsy technologies, however, are limited by the amount of material you collect in the tube of blood,” Love says. “Where this work intercedes is thinking about how to inject something beforehand that would help boost or enhance the amount of signal that is available to collect in the same small sample.”
The body uses two primary strategies to remove circulating DNA from the bloodstream. Enzymes called DNases circulate in the blood and break down DNA that they encounter, while immune cells known as macrophages take up cell-free DNA as blood is filtered through the liver.
The researchers decided to target each of these processes separately. To prevent DNases from breaking down DNA, they designed a monoclonal antibody that binds to circulating DNA and protects it from the enzymes.
“Antibodies are well-established biopharmaceutical modalities, and they’re safe in a number of different disease contexts, including cancer and autoimmune treatments,” Love says. “The idea was, could we use this kind of antibody to help shield the DNA temporarily from degradation by the nucleases that are in circulation? And by doing so, we shift the balance to where the tumor is generating DNA slightly faster than is being degraded, increasing the concentration in a blood draw.”
The other priming agent they developed is a nanoparticle designed to block macrophages from taking up cell-free DNA. These cells have a well-known tendency to eat up synthetic nanoparticles.
“DNA is a biological nanoparticle, and it made sense that immune cells in the liver were probably taking this up just like they do synthetic nanoparticles. And if that were the case, which it turned out to be, then we could use a safe dummy nanoparticle to distract those immune cells and leave the circulating DNA alone so that it could be at a higher concentration,” Bhatia says.
Earlier tumor detection
The researchers tested their priming agents in mice that received transplants of cancer cells that tend to form tumors in the lungs. Two weeks after the cells were transplanted, the researchers showed that these priming agents could boost the amount of circulating tumor DNA recovered in a blood sample by up to 60-fold.
Once the blood sample is taken, it can be run through the same kinds of sequencing tests now used on liquid biopsy samples. These tests can pick out tumor DNA, including specific sequences used to determine the type of tumor and potentially what kinds of treatments would work best.
Early detection of cancer is another promising application for these priming agents. The researchers found that when mice were given the nanoparticle priming agent before blood was drawn, it allowed them to detect circulating tumor DNA in blood of 75 percent of the mice with low cancer burden, while none were detectable without this boost.
“One of the greatest hurdles for cancer liquid biopsy testing has been the scarcity of circulating tumor DNA in a blood sample,” Adalsteinsson says. “It’s thus been encouraging to see the magnitude of the effect we’ve been able to achieve so far and to envision what impact this could have for patients.”
After either of the priming agents are injected, it takes an hour or two for the DNA levels to increase in the bloodstream, and then they return to normal within about 24 hours.
“The ability to get peak activity of these agents within a couple of hours, followed by their rapid clearance, means that someone could go into a doctor’s office, receive an agent like this, and then give their blood for the test itself, all within one visit,” Love says. “This feature bodes well for the potential to translate this concept into clinical use.”
The researchers have launched a company called Amplifyer Bio that plans to further develop the technology, in hopes of advancing to clinical trials.
“A tube of blood is a much more accessible diagnostic than colonoscopy screening or even mammography,” Bhatia says. “Ultimately, if these tools really are predictive, then we should be able to get many more patients into the system who could benefit from cancer interception or better therapy.”
The research was funded by the Koch Institute Support (core) Grant from the National Cancer Institute, the Marble Center for Cancer Nanomedicine, the Gerstner Family Foundation, the Ludwig Center at MIT, the Koch Institute Frontier Research Program via the Casey and Family Foundation, and the Bridge Project, a partnership between the Koch Institute and the Dana-Farber/Harvard Cancer Center.

© Image: MIT News; iStock
Using a virus-like delivery particle made from DNA, researchers from MIT and the Ragon Institute of MGH, MIT, and Harvard have created a vaccine that can induce a strong antibody response against SARS-CoV-2.
The vaccine, which has been tested in mice, consists of a DNA scaffold that carries many copies of a viral antigen. This type of vaccine, known as a particulate vaccine, mimics the structure of a virus. Most previous work on particulate vaccines has relied on protein scaffolds, but the proteins used in those vaccines tend to generate an unnecessary immune response that can distract the immune system from the target.
In the mouse study, the researchers found that the DNA scaffold does not induce an immune response, allowing the immune system to focus its antibody response on the target antigen.
“DNA, we found in this work, does not elicit antibodies that may distract away from the protein of interest,” says Mark Bathe, an MIT professor of biological engineering. “What you can imagine is that your B cells and immune system are being fully trained by that target antigen, and that’s what you want — for your immune system to be laser-focused on the antigen of interest.”
This approach, which strongly stimulates B cells (the cells that produce antibodies), could make it easier to develop vaccines against viruses that have been difficult to target, including HIV and influenza, as well as SARS-CoV-2, the researchers say. Unlike T cells, which are stimulated by other types of vaccines, these B cells can persist for decades, offering long-term protection.
“We’re interested in exploring whether we can teach the immune system to deliver higher levels of immunity against pathogens that resist conventional vaccine approaches, like flu, HIV, and SARS-CoV-2,” says Daniel Lingwood, an associate professor at Harvard Medical School and a principal investigator at the Ragon Institute. “This idea of decoupling the response against the target antigen from the platform itself is a potentially powerful immunological trick that one can now bring to bear to help those immunological targeting decisions move in a direction that is more focused.”
Bathe, Lingwood, and Aaron Schmidt, an associate professor at Harvard Medical School and principal investigator at the Ragon Institute, are the senior authors of the paper, which appears today in Nature Communications. The paper’s lead authors are Eike-Christian Wamhoff, a former MIT postdoc; Larance Ronsard, a Ragon Institute postdoc; Jared Feldman, a former Harvard University graduate student; Grant Knappe, an MIT graduate student; and Blake Hauser, a former Harvard graduate student.
Mimicking viruses
Particulate vaccines usually consist of a protein nanoparticle, similar in structure to a virus, that can carry many copies of a viral antigen. This high density of antigens can lead to a stronger immune response than traditional vaccines because the body sees it as similar to an actual virus. Particulate vaccines have been developed for a handful of pathogens, including hepatitis B and human papillomavirus, and a particulate vaccine for SARS-CoV-2 has been approved for use in South Korea.
These vaccines are especially good at activating B cells, which produce antibodies specific to the vaccine antigen.
“Particulate vaccines are of great interest for many in immunology because they give you robust humoral immunity, which is antibody-based immunity, which is differentiated from the T-cell-based immunity that the mRNA vaccines seem to elicit more strongly,” Bathe says.
A potential drawback to this kind of vaccine, however, is that the proteins used for the scaffold often stimulate the body to produce antibodies targeting the scaffold. This can distract the immune system and prevent it from launching as robust a response as one would like, Bathe says.
“To neutralize the SARS-CoV-2 virus, you want to have a vaccine that generates antibodies toward the receptor binding domain portion of the virus’ spike protein,” he says. “When you display that on a protein-based particle, what happens is your immune system recognizes not only that receptor binding domain protein, but all the other proteins that are irrelevant to the immune response you’re trying to elicit.”
Another potential drawback is that if the same person receives more than one vaccine carried by the same protein scaffold, for example, SARS-CoV-2 and then influenza, their immune system would likely respond right away to the protein scaffold, having already been primed to react to it. This could weaken the immune response to the antigen carried by the second vaccine.
“If you want to apply that protein-based particle to immunize against a different virus like influenza, then your immune system can be addicted to the underlying protein scaffold that it’s already seen and developed an immune response toward,” Bathe says. “That can hypothetically diminish the quality of your antibody response for the actual antigen of interest.”
As an alternative, Bathe’s lab has been developing scaffolds made using DNA origami, a method that offers precise control over the structure of synthetic DNA and allows researchers to attach a variety of molecules, such as viral antigens, at specific locations.
In a 2020 study, Bathe and Darrell Irvine, an MIT professor of biological engineering and of materials science and engineering, showed that a DNA scaffold carrying 30 copies of an HIV antigen could generate a strong antibody response in B cells grown in the lab. This type of structure is optimal for activating B cells because it closely mimics the structure of nano-sized viruses, which display many copies of viral proteins in their surfaces.
“This approach builds off of a fundamental principle in B-cell antigen recognition, which is that if you have an arrayed display of the antigen, that promotes B-cell responses and gives better quantity and quality of antibody output,” Lingwood says.
“Immunologically silent”
In the new study, the researchers swapped in an antigen consisting of the receptor binding protein of the spike protein from the original strain of SARS-CoV-2. When they gave the vaccine to mice, they found that the mice generated high levels of antibodies to the spike protein but did not generate any to the DNA scaffold.
In contrast, a vaccine based on a scaffold protein called ferritin, coated with SARS-CoV-2 antigens, generated many antibodies against ferritin as well as SARS-CoV-2.
“The DNA nanoparticle itself is immunogenically silent,” Lingwood says. “If you use a protein-based platform, you get equally high titer antibody responses to the platform and to the antigen of interest, and that can complicate repeated usage of that platform because you’ll develop high affinity immune memory against it.”
Reducing these off-target effects could also help scientists reach the goal of developing a vaccine that would induce broadly neutralizing antibodies to any variant of SARS-CoV-2, or even to all sarbecoviruses, the subgenus of virus that includes SARS-CoV-2 as well as the viruses that cause SARS and MERS.
To that end, the researchers are now exploring whether a DNA scaffold with many different viral antigens attached could induce broadly neutralizing antibodies against SARS-CoV-2 and related viruses.
The research was primarily funded by the National Institutes of Health, the National Science Foundation, and the Fast Grants program.

© Credit: The Bathe Lab
Tumors constantly shed DNA from dying cells, which briefly circulates in the patient’s bloodstream before it is quickly broken down. Many companies have created blood tests that can pick out this tumor DNA, potentially helping doctors diagnose or monitor cancer or choose a treatment.
The amount of tumor DNA circulating at any given time, however, is extremely small, so it has been challenging to develop tests sensitive enough to pick up that tiny signal. A team of researchers from MIT and the Broad Institute of MIT and Harvard has now come up with a way to significantly boost that signal, by temporarily slowing the clearance of tumor DNA circulating in the bloodstream.
The researchers developed two different types of injectable molecules that they call “priming agents,” which can transiently interfere with the body’s ability to remove circulating tumor DNA from the bloodstream. In a study of mice, they showed that these agents could boost DNA levels enough that the percentage of detectable early-stage lung metastases leapt from less than 10 percent to above 75 percent.
This approach could enable not only earlier diagnosis of cancer, but also more sensitive detection of tumor mutations that could be used to guide treatment. It could also help improve detection of cancer recurrence.
“You can give one of these agents an hour before the blood draw, and it makes things visible that previously wouldn’t have been. The implication is that we should be able to give everybody who’s doing liquid biopsies, for any purpose, more molecules to work with,” says Sangeeta Bhatia, the John and Dorothy Wilson Professor of Health Sciences and Technology and of Electrical Engineering and Computer Science at MIT, and a member of MIT’s Koch Institute for Integrative Cancer Research and the Institute for Medical Engineering and Science.
Bhatia is one of the senior authors of the new study, along with J. Christopher Love, the Raymond A. and Helen E. St. Laurent Professor of Chemical Engineering at MIT and a member of the Koch Institute and the Ragon Institute of MGH, MIT, and Harvard and Viktor Adalsteinsson, director of the Gerstner Center for Cancer Diagnostics at the Broad Institute.
Carmen Martin-Alonso PhD ’23, MIT and Broad Institute postdoc Shervin Tabrizi, and Broad Institute scientist Kan Xiong are the lead authors of the paper, which appears today in Science.
Better biopsies
Liquid biopsies, which enable detection of small quantities of DNA in blood samples, are now used in many cancer patients to identify mutations that could help guide treatment. With greater sensitivity, however, these tests could become useful for far more patients. Most efforts to improve the sensitivity of liquid biopsies have focused on developing new sequencing technologies to use after the blood is drawn.
While brainstorming ways to make liquid biopsies more informative, Bhatia, Love, Adalsteinsson, and their trainees came up with the idea of trying to increase the amount of DNA in a patient’s bloodstream before the sample is taken.
“A tumor is always creating new cell-free DNA, and that’s the signal that we’re attempting to detect in the blood draw. Existing liquid biopsy technologies, however, are limited by the amount of material you collect in the tube of blood,” Love says. “Where this work intercedes is thinking about how to inject something beforehand that would help boost or enhance the amount of signal that is available to collect in the same small sample.”
The body uses two primary strategies to remove circulating DNA from the bloodstream. Enzymes called DNases circulate in the blood and break down DNA that they encounter, while immune cells known as macrophages take up cell-free DNA as blood is filtered through the liver.
The researchers decided to target each of these processes separately. To prevent DNases from breaking down DNA, they designed a monoclonal antibody that binds to circulating DNA and protects it from the enzymes.
“Antibodies are well-established biopharmaceutical modalities, and they’re safe in a number of different disease contexts, including cancer and autoimmune treatments,” Love says. “The idea was, could we use this kind of antibody to help shield the DNA temporarily from degradation by the nucleases that are in circulation? And by doing so, we shift the balance to where the tumor is generating DNA slightly faster than is being degraded, increasing the concentration in a blood draw.”
The other priming agent they developed is a nanoparticle designed to block macrophages from taking up cell-free DNA. These cells have a well-known tendency to eat up synthetic nanoparticles.
“DNA is a biological nanoparticle, and it made sense that immune cells in the liver were probably taking this up just like they do synthetic nanoparticles. And if that were the case, which it turned out to be, then we could use a safe dummy nanoparticle to distract those immune cells and leave the circulating DNA alone so that it could be at a higher concentration,” Bhatia says.
Earlier tumor detection
The researchers tested their priming agents in mice that received transplants of cancer cells that tend to form tumors in the lungs. Two weeks after the cells were transplanted, the researchers showed that these priming agents could boost the amount of circulating tumor DNA recovered in a blood sample by up to 60-fold.
Once the blood sample is taken, it can be run through the same kinds of sequencing tests now used on liquid biopsy samples. These tests can pick out tumor DNA, including specific sequences used to determine the type of tumor and potentially what kinds of treatments would work best.
Early detection of cancer is another promising application for these priming agents. The researchers found that when mice were given the nanoparticle priming agent before blood was drawn, it allowed them to detect circulating tumor DNA in blood of 75 percent of the mice with low cancer burden, while none were detectable without this boost.
“One of the greatest hurdles for cancer liquid biopsy testing has been the scarcity of circulating tumor DNA in a blood sample,” Adalsteinsson says. “It’s thus been encouraging to see the magnitude of the effect we’ve been able to achieve so far and to envision what impact this could have for patients.”
After either of the priming agents are injected, it takes an hour or two for the DNA levels to increase in the bloodstream, and then they return to normal within about 24 hours.
“The ability to get peak activity of these agents within a couple of hours, followed by their rapid clearance, means that someone could go into a doctor’s office, receive an agent like this, and then give their blood for the test itself, all within one visit,” Love says. “This feature bodes well for the potential to translate this concept into clinical use.”
The researchers have launched a company called Amplifyer Bio that plans to further develop the technology, in hopes of advancing to clinical trials.
“A tube of blood is a much more accessible diagnostic than colonoscopy screening or even mammography,” Bhatia says. “Ultimately, if these tools really are predictive, then we should be able to get many more patients into the system who could benefit from cancer interception or better therapy.”
The research was funded by the Koch Institute Support (core) Grant from the National Cancer Institute, the Marble Center for Cancer Nanomedicine, the Gerstner Family Foundation, the Ludwig Center at MIT, the Koch Institute Frontier Research Program via the Casey and Family Foundation, and the Bridge Project, a partnership between the Koch Institute and the Dana-Farber/Harvard Cancer Center.

© Image: MIT News; iStock
AMD GPUOpen - Graphics and game developer resources
A repository of AMD Instruction Set Architecture (ISA) and Micro Engine Scheduler (MES) firmware documentation
The post AMD GPU architecture programming documentation appeared first on AMD GPUOpen.

The Sonic the Hedgehog franchise has seen significantly more television and film adaptations than most video game series can claim. While it’s having its most widespread moment ever with its current live-action movie franchise, the blue blur has been starring in animated series for over 30 years. Each of these…

Much like a Sonic the Hedgehog level, Sega’s platforming franchise has had its ups, downs, loop-de-loops, and bottomless chasms. With the Knuckles show out, a third live-action movie premiering in December, and Sonic X Shadow Generations coming out this year, what better time to look back at the Sonic franchise’s best…

Looking at the original trailer for the live-action Sonic the Hedgehog movie is like looking into a portal to a different timeline, one where the film itself likely doesn’t herald the arrival of a massively successful film franchise that eventually spawns a bad Paramount+ spin-off. At best, it results in a Morbius-leve…

Enlarge / A decorative ring made from carved stone is embedded in the wall of a ballcourt in the ancient Maya city of Chichen Itza. (credit: Kåre Thor Olsen/CC BY-SA 3.0)
It's well-known that the ancient Maya had their own version of ball games, which were played with a rubber ball on stone courts. Such games served not just as athletic events but also religious ones that often involved ritual sacrifices. Archaeologists have now found evidence that the Maya may have blessed newly constructed ball courts in rituals involving plants with hallucinogenic properties, according to a new paper published in the journal PLoS ONE.
“When they erected a new building, they asked the goodwill of the gods to protect the people inhabiting it,” said co-author David Lentz of the University of Cincinnati. “Some people call it an ensouling ritual, to get a blessing from and appease the gods.” Lentz and his team previously used genetic and pollen analyses of the wild and cultivated plants found in the ancient Maya city Yaxnohcah in what is now Mexico’s Yucatan Peninsula, revealing evidence of sustainable agriculture and forestry spanning a millennia.
As we've reported previously, there is ample evidence that humans in many cultures throughout history used various hallucinogenic substances in religious ceremonies or shamanic rituals. That includes not just ancient Egypt but also ancient Greek, Vedic, Maya, Inca, and Aztec cultures. The Urarina people who live in the Peruvian Amazon Basin still use a psychoactive brew called ayahuasca in their rituals, and Westerners seeking their own brand of enlightenment have also been known to participate.
Using a virus-like delivery particle made from DNA, researchers from MIT and the Ragon Institute of MGH, MIT, and Harvard have created a vaccine that can induce a strong antibody response against SARS-CoV-2.
The vaccine, which has been tested in mice, consists of a DNA scaffold that carries many copies of a viral antigen. This type of vaccine, known as a particulate vaccine, mimics the structure of a virus. Most previous work on particulate vaccines has relied on protein scaffolds, but the proteins used in those vaccines tend to generate an unnecessary immune response that can distract the immune system from the target.
In the mouse study, the researchers found that the DNA scaffold does not induce an immune response, allowing the immune system to focus its antibody response on the target antigen.
“DNA, we found in this work, does not elicit antibodies that may distract away from the protein of interest,” says Mark Bathe, an MIT professor of biological engineering. “What you can imagine is that your B cells and immune system are being fully trained by that target antigen, and that’s what you want — for your immune system to be laser-focused on the antigen of interest.”
This approach, which strongly stimulates B cells (the cells that produce antibodies), could make it easier to develop vaccines against viruses that have been difficult to target, including HIV and influenza, as well as SARS-CoV-2, the researchers say. Unlike T cells, which are stimulated by other types of vaccines, these B cells can persist for decades, offering long-term protection.
“We’re interested in exploring whether we can teach the immune system to deliver higher levels of immunity against pathogens that resist conventional vaccine approaches, like flu, HIV, and SARS-CoV-2,” says Daniel Lingwood, an associate professor at Harvard Medical School and a principal investigator at the Ragon Institute. “This idea of decoupling the response against the target antigen from the platform itself is a potentially powerful immunological trick that one can now bring to bear to help those immunological targeting decisions move in a direction that is more focused.”
Bathe, Lingwood, and Aaron Schmidt, an associate professor at Harvard Medical School and principal investigator at the Ragon Institute, are the senior authors of the paper, which appears today in Nature Communications. The paper’s lead authors are Eike-Christian Wamhoff, a former MIT postdoc; Larance Ronsard, a Ragon Institute postdoc; Jared Feldman, a former Harvard University graduate student; Grant Knappe, an MIT graduate student; and Blake Hauser, a former Harvard graduate student.
Mimicking viruses
Particulate vaccines usually consist of a protein nanoparticle, similar in structure to a virus, that can carry many copies of a viral antigen. This high density of antigens can lead to a stronger immune response than traditional vaccines because the body sees it as similar to an actual virus. Particulate vaccines have been developed for a handful of pathogens, including hepatitis B and human papillomavirus, and a particulate vaccine for SARS-CoV-2 has been approved for use in South Korea.
These vaccines are especially good at activating B cells, which produce antibodies specific to the vaccine antigen.
“Particulate vaccines are of great interest for many in immunology because they give you robust humoral immunity, which is antibody-based immunity, which is differentiated from the T-cell-based immunity that the mRNA vaccines seem to elicit more strongly,” Bathe says.
A potential drawback to this kind of vaccine, however, is that the proteins used for the scaffold often stimulate the body to produce antibodies targeting the scaffold. This can distract the immune system and prevent it from launching as robust a response as one would like, Bathe says.
“To neutralize the SARS-CoV-2 virus, you want to have a vaccine that generates antibodies toward the receptor binding domain portion of the virus’ spike protein,” he says. “When you display that on a protein-based particle, what happens is your immune system recognizes not only that receptor binding domain protein, but all the other proteins that are irrelevant to the immune response you’re trying to elicit.”
Another potential drawback is that if the same person receives more than one vaccine carried by the same protein scaffold, for example, SARS-CoV-2 and then influenza, their immune system would likely respond right away to the protein scaffold, having already been primed to react to it. This could weaken the immune response to the antigen carried by the second vaccine.
“If you want to apply that protein-based particle to immunize against a different virus like influenza, then your immune system can be addicted to the underlying protein scaffold that it’s already seen and developed an immune response toward,” Bathe says. “That can hypothetically diminish the quality of your antibody response for the actual antigen of interest.”
As an alternative, Bathe’s lab has been developing scaffolds made using DNA origami, a method that offers precise control over the structure of synthetic DNA and allows researchers to attach a variety of molecules, such as viral antigens, at specific locations.
In a 2020 study, Bathe and Darrell Irvine, an MIT professor of biological engineering and of materials science and engineering, showed that a DNA scaffold carrying 30 copies of an HIV antigen could generate a strong antibody response in B cells grown in the lab. This type of structure is optimal for activating B cells because it closely mimics the structure of nano-sized viruses, which display many copies of viral proteins in their surfaces.
“This approach builds off of a fundamental principle in B-cell antigen recognition, which is that if you have an arrayed display of the antigen, that promotes B-cell responses and gives better quantity and quality of antibody output,” Lingwood says.
“Immunologically silent”
In the new study, the researchers swapped in an antigen consisting of the receptor binding protein of the spike protein from the original strain of SARS-CoV-2. When they gave the vaccine to mice, they found that the mice generated high levels of antibodies to the spike protein but did not generate any to the DNA scaffold.
In contrast, a vaccine based on a scaffold protein called ferritin, coated with SARS-CoV-2 antigens, generated many antibodies against ferritin as well as SARS-CoV-2.
“The DNA nanoparticle itself is immunogenically silent,” Lingwood says. “If you use a protein-based platform, you get equally high titer antibody responses to the platform and to the antigen of interest, and that can complicate repeated usage of that platform because you’ll develop high affinity immune memory against it.”
Reducing these off-target effects could also help scientists reach the goal of developing a vaccine that would induce broadly neutralizing antibodies to any variant of SARS-CoV-2, or even to all sarbecoviruses, the subgenus of virus that includes SARS-CoV-2 as well as the viruses that cause SARS and MERS.
To that end, the researchers are now exploring whether a DNA scaffold with many different viral antigens attached could induce broadly neutralizing antibodies against SARS-CoV-2 and related viruses.
The research was primarily funded by the National Institutes of Health, the National Science Foundation, and the Fast Grants program.

© Credit: The Bathe Lab
Tumors constantly shed DNA from dying cells, which briefly circulates in the patient’s bloodstream before it is quickly broken down. Many companies have created blood tests that can pick out this tumor DNA, potentially helping doctors diagnose or monitor cancer or choose a treatment.
The amount of tumor DNA circulating at any given time, however, is extremely small, so it has been challenging to develop tests sensitive enough to pick up that tiny signal. A team of researchers from MIT and the Broad Institute of MIT and Harvard has now come up with a way to significantly boost that signal, by temporarily slowing the clearance of tumor DNA circulating in the bloodstream.
The researchers developed two different types of injectable molecules that they call “priming agents,” which can transiently interfere with the body’s ability to remove circulating tumor DNA from the bloodstream. In a study of mice, they showed that these agents could boost DNA levels enough that the percentage of detectable early-stage lung metastases leapt from less than 10 percent to above 75 percent.
This approach could enable not only earlier diagnosis of cancer, but also more sensitive detection of tumor mutations that could be used to guide treatment. It could also help improve detection of cancer recurrence.
“You can give one of these agents an hour before the blood draw, and it makes things visible that previously wouldn’t have been. The implication is that we should be able to give everybody who’s doing liquid biopsies, for any purpose, more molecules to work with,” says Sangeeta Bhatia, the John and Dorothy Wilson Professor of Health Sciences and Technology and of Electrical Engineering and Computer Science at MIT, and a member of MIT’s Koch Institute for Integrative Cancer Research and the Institute for Medical Engineering and Science.
Bhatia is one of the senior authors of the new study, along with J. Christopher Love, the Raymond A. and Helen E. St. Laurent Professor of Chemical Engineering at MIT and a member of the Koch Institute and the Ragon Institute of MGH, MIT, and Harvard and Viktor Adalsteinsson, director of the Gerstner Center for Cancer Diagnostics at the Broad Institute.
Carmen Martin-Alonso PhD ’23, MIT and Broad Institute postdoc Shervin Tabrizi, and Broad Institute scientist Kan Xiong are the lead authors of the paper, which appears today in Science.
Better biopsies
Liquid biopsies, which enable detection of small quantities of DNA in blood samples, are now used in many cancer patients to identify mutations that could help guide treatment. With greater sensitivity, however, these tests could become useful for far more patients. Most efforts to improve the sensitivity of liquid biopsies have focused on developing new sequencing technologies to use after the blood is drawn.
While brainstorming ways to make liquid biopsies more informative, Bhatia, Love, Adalsteinsson, and their trainees came up with the idea of trying to increase the amount of DNA in a patient’s bloodstream before the sample is taken.
“A tumor is always creating new cell-free DNA, and that’s the signal that we’re attempting to detect in the blood draw. Existing liquid biopsy technologies, however, are limited by the amount of material you collect in the tube of blood,” Love says. “Where this work intercedes is thinking about how to inject something beforehand that would help boost or enhance the amount of signal that is available to collect in the same small sample.”
The body uses two primary strategies to remove circulating DNA from the bloodstream. Enzymes called DNases circulate in the blood and break down DNA that they encounter, while immune cells known as macrophages take up cell-free DNA as blood is filtered through the liver.
The researchers decided to target each of these processes separately. To prevent DNases from breaking down DNA, they designed a monoclonal antibody that binds to circulating DNA and protects it from the enzymes.
“Antibodies are well-established biopharmaceutical modalities, and they’re safe in a number of different disease contexts, including cancer and autoimmune treatments,” Love says. “The idea was, could we use this kind of antibody to help shield the DNA temporarily from degradation by the nucleases that are in circulation? And by doing so, we shift the balance to where the tumor is generating DNA slightly faster than is being degraded, increasing the concentration in a blood draw.”
The other priming agent they developed is a nanoparticle designed to block macrophages from taking up cell-free DNA. These cells have a well-known tendency to eat up synthetic nanoparticles.
“DNA is a biological nanoparticle, and it made sense that immune cells in the liver were probably taking this up just like they do synthetic nanoparticles. And if that were the case, which it turned out to be, then we could use a safe dummy nanoparticle to distract those immune cells and leave the circulating DNA alone so that it could be at a higher concentration,” Bhatia says.
Earlier tumor detection
The researchers tested their priming agents in mice that received transplants of cancer cells that tend to form tumors in the lungs. Two weeks after the cells were transplanted, the researchers showed that these priming agents could boost the amount of circulating tumor DNA recovered in a blood sample by up to 60-fold.
Once the blood sample is taken, it can be run through the same kinds of sequencing tests now used on liquid biopsy samples. These tests can pick out tumor DNA, including specific sequences used to determine the type of tumor and potentially what kinds of treatments would work best.
Early detection of cancer is another promising application for these priming agents. The researchers found that when mice were given the nanoparticle priming agent before blood was drawn, it allowed them to detect circulating tumor DNA in blood of 75 percent of the mice with low cancer burden, while none were detectable without this boost.
“One of the greatest hurdles for cancer liquid biopsy testing has been the scarcity of circulating tumor DNA in a blood sample,” Adalsteinsson says. “It’s thus been encouraging to see the magnitude of the effect we’ve been able to achieve so far and to envision what impact this could have for patients.”
After either of the priming agents are injected, it takes an hour or two for the DNA levels to increase in the bloodstream, and then they return to normal within about 24 hours.
“The ability to get peak activity of these agents within a couple of hours, followed by their rapid clearance, means that someone could go into a doctor’s office, receive an agent like this, and then give their blood for the test itself, all within one visit,” Love says. “This feature bodes well for the potential to translate this concept into clinical use.”
The researchers have launched a company called Amplifyer Bio that plans to further develop the technology, in hopes of advancing to clinical trials.
“A tube of blood is a much more accessible diagnostic than colonoscopy screening or even mammography,” Bhatia says. “Ultimately, if these tools really are predictive, then we should be able to get many more patients into the system who could benefit from cancer interception or better therapy.”
The research was funded by the Koch Institute Support (core) Grant from the National Cancer Institute, the Marble Center for Cancer Nanomedicine, the Gerstner Family Foundation, the Ludwig Center at MIT, the Koch Institute Frontier Research Program via the Casey and Family Foundation, and the Bridge Project, a partnership between the Koch Institute and the Dana-Farber/Harvard Cancer Center.

© Image: MIT News; iStock

I had a lot of expectations for the Knuckles live-action mini-series that, it turns out, might have been a bit too lofty. But I don’t think the expectation that the titular echidna voiced by Idris Elbawould be in a show with his name at the top was that outrageous. I’m sad to report that Knuckles is only around for a…

It’s a good thing Knuckles, the six-episode Paramount+ mini-series is coming out during a fever pitch of hype for Sonic the Hedgehog 3. The series debuts on the streaming service on April 26, during a time when fans are dying to get back into the live-action universe off of announcements like Keanu Reeves playing the…
Using a virus-like delivery particle made from DNA, researchers from MIT and the Ragon Institute of MGH, MIT, and Harvard have created a vaccine that can induce a strong antibody response against SARS-CoV-2.
The vaccine, which has been tested in mice, consists of a DNA scaffold that carries many copies of a viral antigen. This type of vaccine, known as a particulate vaccine, mimics the structure of a virus. Most previous work on particulate vaccines has relied on protein scaffolds, but the proteins used in those vaccines tend to generate an unnecessary immune response that can distract the immune system from the target.
In the mouse study, the researchers found that the DNA scaffold does not induce an immune response, allowing the immune system to focus its antibody response on the target antigen.
“DNA, we found in this work, does not elicit antibodies that may distract away from the protein of interest,” says Mark Bathe, an MIT professor of biological engineering. “What you can imagine is that your B cells and immune system are being fully trained by that target antigen, and that’s what you want — for your immune system to be laser-focused on the antigen of interest.”
This approach, which strongly stimulates B cells (the cells that produce antibodies), could make it easier to develop vaccines against viruses that have been difficult to target, including HIV and influenza, as well as SARS-CoV-2, the researchers say. Unlike T cells, which are stimulated by other types of vaccines, these B cells can persist for decades, offering long-term protection.
“We’re interested in exploring whether we can teach the immune system to deliver higher levels of immunity against pathogens that resist conventional vaccine approaches, like flu, HIV, and SARS-CoV-2,” says Daniel Lingwood, an associate professor at Harvard Medical School and a principal investigator at the Ragon Institute. “This idea of decoupling the response against the target antigen from the platform itself is a potentially powerful immunological trick that one can now bring to bear to help those immunological targeting decisions move in a direction that is more focused.”
Bathe, Lingwood, and Aaron Schmidt, an associate professor at Harvard Medical School and principal investigator at the Ragon Institute, are the senior authors of the paper, which appears today in Nature Communications. The paper’s lead authors are Eike-Christian Wamhoff, a former MIT postdoc; Larance Ronsard, a Ragon Institute postdoc; Jared Feldman, a former Harvard University graduate student; Grant Knappe, an MIT graduate student; and Blake Hauser, a former Harvard graduate student.
Mimicking viruses
Particulate vaccines usually consist of a protein nanoparticle, similar in structure to a virus, that can carry many copies of a viral antigen. This high density of antigens can lead to a stronger immune response than traditional vaccines because the body sees it as similar to an actual virus. Particulate vaccines have been developed for a handful of pathogens, including hepatitis B and human papillomavirus, and a particulate vaccine for SARS-CoV-2 has been approved for use in South Korea.
These vaccines are especially good at activating B cells, which produce antibodies specific to the vaccine antigen.
“Particulate vaccines are of great interest for many in immunology because they give you robust humoral immunity, which is antibody-based immunity, which is differentiated from the T-cell-based immunity that the mRNA vaccines seem to elicit more strongly,” Bathe says.
A potential drawback to this kind of vaccine, however, is that the proteins used for the scaffold often stimulate the body to produce antibodies targeting the scaffold. This can distract the immune system and prevent it from launching as robust a response as one would like, Bathe says.
“To neutralize the SARS-CoV-2 virus, you want to have a vaccine that generates antibodies toward the receptor binding domain portion of the virus’ spike protein,” he says. “When you display that on a protein-based particle, what happens is your immune system recognizes not only that receptor binding domain protein, but all the other proteins that are irrelevant to the immune response you’re trying to elicit.”
Another potential drawback is that if the same person receives more than one vaccine carried by the same protein scaffold, for example, SARS-CoV-2 and then influenza, their immune system would likely respond right away to the protein scaffold, having already been primed to react to it. This could weaken the immune response to the antigen carried by the second vaccine.
“If you want to apply that protein-based particle to immunize against a different virus like influenza, then your immune system can be addicted to the underlying protein scaffold that it’s already seen and developed an immune response toward,” Bathe says. “That can hypothetically diminish the quality of your antibody response for the actual antigen of interest.”
As an alternative, Bathe’s lab has been developing scaffolds made using DNA origami, a method that offers precise control over the structure of synthetic DNA and allows researchers to attach a variety of molecules, such as viral antigens, at specific locations.
In a 2020 study, Bathe and Darrell Irvine, an MIT professor of biological engineering and of materials science and engineering, showed that a DNA scaffold carrying 30 copies of an HIV antigen could generate a strong antibody response in B cells grown in the lab. This type of structure is optimal for activating B cells because it closely mimics the structure of nano-sized viruses, which display many copies of viral proteins in their surfaces.
“This approach builds off of a fundamental principle in B-cell antigen recognition, which is that if you have an arrayed display of the antigen, that promotes B-cell responses and gives better quantity and quality of antibody output,” Lingwood says.
“Immunologically silent”
In the new study, the researchers swapped in an antigen consisting of the receptor binding protein of the spike protein from the original strain of SARS-CoV-2. When they gave the vaccine to mice, they found that the mice generated high levels of antibodies to the spike protein but did not generate any to the DNA scaffold.
In contrast, a vaccine based on a scaffold protein called ferritin, coated with SARS-CoV-2 antigens, generated many antibodies against ferritin as well as SARS-CoV-2.
“The DNA nanoparticle itself is immunogenically silent,” Lingwood says. “If you use a protein-based platform, you get equally high titer antibody responses to the platform and to the antigen of interest, and that can complicate repeated usage of that platform because you’ll develop high affinity immune memory against it.”
Reducing these off-target effects could also help scientists reach the goal of developing a vaccine that would induce broadly neutralizing antibodies to any variant of SARS-CoV-2, or even to all sarbecoviruses, the subgenus of virus that includes SARS-CoV-2 as well as the viruses that cause SARS and MERS.
To that end, the researchers are now exploring whether a DNA scaffold with many different viral antigens attached could induce broadly neutralizing antibodies against SARS-CoV-2 and related viruses.
The research was primarily funded by the National Institutes of Health, the National Science Foundation, and the Fast Grants program.

© Credit: The Bathe Lab
Tumors constantly shed DNA from dying cells, which briefly circulates in the patient’s bloodstream before it is quickly broken down. Many companies have created blood tests that can pick out this tumor DNA, potentially helping doctors diagnose or monitor cancer or choose a treatment.
The amount of tumor DNA circulating at any given time, however, is extremely small, so it has been challenging to develop tests sensitive enough to pick up that tiny signal. A team of researchers from MIT and the Broad Institute of MIT and Harvard has now come up with a way to significantly boost that signal, by temporarily slowing the clearance of tumor DNA circulating in the bloodstream.
The researchers developed two different types of injectable molecules that they call “priming agents,” which can transiently interfere with the body’s ability to remove circulating tumor DNA from the bloodstream. In a study of mice, they showed that these agents could boost DNA levels enough that the percentage of detectable early-stage lung metastases leapt from less than 10 percent to above 75 percent.
This approach could enable not only earlier diagnosis of cancer, but also more sensitive detection of tumor mutations that could be used to guide treatment. It could also help improve detection of cancer recurrence.
“You can give one of these agents an hour before the blood draw, and it makes things visible that previously wouldn’t have been. The implication is that we should be able to give everybody who’s doing liquid biopsies, for any purpose, more molecules to work with,” says Sangeeta Bhatia, the John and Dorothy Wilson Professor of Health Sciences and Technology and of Electrical Engineering and Computer Science at MIT, and a member of MIT’s Koch Institute for Integrative Cancer Research and the Institute for Medical Engineering and Science.
Bhatia is one of the senior authors of the new study, along with J. Christopher Love, the Raymond A. and Helen E. St. Laurent Professor of Chemical Engineering at MIT and a member of the Koch Institute and the Ragon Institute of MGH, MIT, and Harvard and Viktor Adalsteinsson, director of the Gerstner Center for Cancer Diagnostics at the Broad Institute.
Carmen Martin-Alonso PhD ’23, MIT and Broad Institute postdoc Shervin Tabrizi, and Broad Institute scientist Kan Xiong are the lead authors of the paper, which appears today in Science.
Better biopsies
Liquid biopsies, which enable detection of small quantities of DNA in blood samples, are now used in many cancer patients to identify mutations that could help guide treatment. With greater sensitivity, however, these tests could become useful for far more patients. Most efforts to improve the sensitivity of liquid biopsies have focused on developing new sequencing technologies to use after the blood is drawn.
While brainstorming ways to make liquid biopsies more informative, Bhatia, Love, Adalsteinsson, and their trainees came up with the idea of trying to increase the amount of DNA in a patient’s bloodstream before the sample is taken.
“A tumor is always creating new cell-free DNA, and that’s the signal that we’re attempting to detect in the blood draw. Existing liquid biopsy technologies, however, are limited by the amount of material you collect in the tube of blood,” Love says. “Where this work intercedes is thinking about how to inject something beforehand that would help boost or enhance the amount of signal that is available to collect in the same small sample.”
The body uses two primary strategies to remove circulating DNA from the bloodstream. Enzymes called DNases circulate in the blood and break down DNA that they encounter, while immune cells known as macrophages take up cell-free DNA as blood is filtered through the liver.
The researchers decided to target each of these processes separately. To prevent DNases from breaking down DNA, they designed a monoclonal antibody that binds to circulating DNA and protects it from the enzymes.
“Antibodies are well-established biopharmaceutical modalities, and they’re safe in a number of different disease contexts, including cancer and autoimmune treatments,” Love says. “The idea was, could we use this kind of antibody to help shield the DNA temporarily from degradation by the nucleases that are in circulation? And by doing so, we shift the balance to where the tumor is generating DNA slightly faster than is being degraded, increasing the concentration in a blood draw.”
The other priming agent they developed is a nanoparticle designed to block macrophages from taking up cell-free DNA. These cells have a well-known tendency to eat up synthetic nanoparticles.
“DNA is a biological nanoparticle, and it made sense that immune cells in the liver were probably taking this up just like they do synthetic nanoparticles. And if that were the case, which it turned out to be, then we could use a safe dummy nanoparticle to distract those immune cells and leave the circulating DNA alone so that it could be at a higher concentration,” Bhatia says.
Earlier tumor detection
The researchers tested their priming agents in mice that received transplants of cancer cells that tend to form tumors in the lungs. Two weeks after the cells were transplanted, the researchers showed that these priming agents could boost the amount of circulating tumor DNA recovered in a blood sample by up to 60-fold.
Once the blood sample is taken, it can be run through the same kinds of sequencing tests now used on liquid biopsy samples. These tests can pick out tumor DNA, including specific sequences used to determine the type of tumor and potentially what kinds of treatments would work best.
Early detection of cancer is another promising application for these priming agents. The researchers found that when mice were given the nanoparticle priming agent before blood was drawn, it allowed them to detect circulating tumor DNA in blood of 75 percent of the mice with low cancer burden, while none were detectable without this boost.
“One of the greatest hurdles for cancer liquid biopsy testing has been the scarcity of circulating tumor DNA in a blood sample,” Adalsteinsson says. “It’s thus been encouraging to see the magnitude of the effect we’ve been able to achieve so far and to envision what impact this could have for patients.”
After either of the priming agents are injected, it takes an hour or two for the DNA levels to increase in the bloodstream, and then they return to normal within about 24 hours.
“The ability to get peak activity of these agents within a couple of hours, followed by their rapid clearance, means that someone could go into a doctor’s office, receive an agent like this, and then give their blood for the test itself, all within one visit,” Love says. “This feature bodes well for the potential to translate this concept into clinical use.”
The researchers have launched a company called Amplifyer Bio that plans to further develop the technology, in hopes of advancing to clinical trials.
“A tube of blood is a much more accessible diagnostic than colonoscopy screening or even mammography,” Bhatia says. “Ultimately, if these tools really are predictive, then we should be able to get many more patients into the system who could benefit from cancer interception or better therapy.”
The research was funded by the Koch Institute Support (core) Grant from the National Cancer Institute, the Marble Center for Cancer Nanomedicine, the Gerstner Family Foundation, the Ludwig Center at MIT, the Koch Institute Frontier Research Program via the Casey and Family Foundation, and the Bridge Project, a partnership between the Koch Institute and the Dana-Farber/Harvard Cancer Center.

© Image: MIT News; iStock
AMD GPUOpen - Graphics and game developer resources
The fourth post in our mesh shaders series takes a look at the specific example of rendering detailed vegetation.
The post Procedural grass rendering – Mesh shaders on AMD RDNA™ graphics cards appeared first on AMD GPUOpen.
AMD GPUOpen - Graphics and game developer resources
The third post in our mesh shaders series covers how to use mesh shaders to simplify font rendering.
The post Font and vector-art rendering with mesh shaders – Mesh shaders on AMD RDNA™ graphics cards appeared first on AMD GPUOpen.
Using a virus-like delivery particle made from DNA, researchers from MIT and the Ragon Institute of MGH, MIT, and Harvard have created a vaccine that can induce a strong antibody response against SARS-CoV-2.
The vaccine, which has been tested in mice, consists of a DNA scaffold that carries many copies of a viral antigen. This type of vaccine, known as a particulate vaccine, mimics the structure of a virus. Most previous work on particulate vaccines has relied on protein scaffolds, but the proteins used in those vaccines tend to generate an unnecessary immune response that can distract the immune system from the target.
In the mouse study, the researchers found that the DNA scaffold does not induce an immune response, allowing the immune system to focus its antibody response on the target antigen.
“DNA, we found in this work, does not elicit antibodies that may distract away from the protein of interest,” says Mark Bathe, an MIT professor of biological engineering. “What you can imagine is that your B cells and immune system are being fully trained by that target antigen, and that’s what you want — for your immune system to be laser-focused on the antigen of interest.”
This approach, which strongly stimulates B cells (the cells that produce antibodies), could make it easier to develop vaccines against viruses that have been difficult to target, including HIV and influenza, as well as SARS-CoV-2, the researchers say. Unlike T cells, which are stimulated by other types of vaccines, these B cells can persist for decades, offering long-term protection.
“We’re interested in exploring whether we can teach the immune system to deliver higher levels of immunity against pathogens that resist conventional vaccine approaches, like flu, HIV, and SARS-CoV-2,” says Daniel Lingwood, an associate professor at Harvard Medical School and a principal investigator at the Ragon Institute. “This idea of decoupling the response against the target antigen from the platform itself is a potentially powerful immunological trick that one can now bring to bear to help those immunological targeting decisions move in a direction that is more focused.”
Bathe, Lingwood, and Aaron Schmidt, an associate professor at Harvard Medical School and principal investigator at the Ragon Institute, are the senior authors of the paper, which appears today in Nature Communications. The paper’s lead authors are Eike-Christian Wamhoff, a former MIT postdoc; Larance Ronsard, a Ragon Institute postdoc; Jared Feldman, a former Harvard University graduate student; Grant Knappe, an MIT graduate student; and Blake Hauser, a former Harvard graduate student.
Mimicking viruses
Particulate vaccines usually consist of a protein nanoparticle, similar in structure to a virus, that can carry many copies of a viral antigen. This high density of antigens can lead to a stronger immune response than traditional vaccines because the body sees it as similar to an actual virus. Particulate vaccines have been developed for a handful of pathogens, including hepatitis B and human papillomavirus, and a particulate vaccine for SARS-CoV-2 has been approved for use in South Korea.
These vaccines are especially good at activating B cells, which produce antibodies specific to the vaccine antigen.
“Particulate vaccines are of great interest for many in immunology because they give you robust humoral immunity, which is antibody-based immunity, which is differentiated from the T-cell-based immunity that the mRNA vaccines seem to elicit more strongly,” Bathe says.
A potential drawback to this kind of vaccine, however, is that the proteins used for the scaffold often stimulate the body to produce antibodies targeting the scaffold. This can distract the immune system and prevent it from launching as robust a response as one would like, Bathe says.
“To neutralize the SARS-CoV-2 virus, you want to have a vaccine that generates antibodies toward the receptor binding domain portion of the virus’ spike protein,” he says. “When you display that on a protein-based particle, what happens is your immune system recognizes not only that receptor binding domain protein, but all the other proteins that are irrelevant to the immune response you’re trying to elicit.”
Another potential drawback is that if the same person receives more than one vaccine carried by the same protein scaffold, for example, SARS-CoV-2 and then influenza, their immune system would likely respond right away to the protein scaffold, having already been primed to react to it. This could weaken the immune response to the antigen carried by the second vaccine.
“If you want to apply that protein-based particle to immunize against a different virus like influenza, then your immune system can be addicted to the underlying protein scaffold that it’s already seen and developed an immune response toward,” Bathe says. “That can hypothetically diminish the quality of your antibody response for the actual antigen of interest.”
As an alternative, Bathe’s lab has been developing scaffolds made using DNA origami, a method that offers precise control over the structure of synthetic DNA and allows researchers to attach a variety of molecules, such as viral antigens, at specific locations.
In a 2020 study, Bathe and Darrell Irvine, an MIT professor of biological engineering and of materials science and engineering, showed that a DNA scaffold carrying 30 copies of an HIV antigen could generate a strong antibody response in B cells grown in the lab. This type of structure is optimal for activating B cells because it closely mimics the structure of nano-sized viruses, which display many copies of viral proteins in their surfaces.
“This approach builds off of a fundamental principle in B-cell antigen recognition, which is that if you have an arrayed display of the antigen, that promotes B-cell responses and gives better quantity and quality of antibody output,” Lingwood says.
“Immunologically silent”
In the new study, the researchers swapped in an antigen consisting of the receptor binding protein of the spike protein from the original strain of SARS-CoV-2. When they gave the vaccine to mice, they found that the mice generated high levels of antibodies to the spike protein but did not generate any to the DNA scaffold.
In contrast, a vaccine based on a scaffold protein called ferritin, coated with SARS-CoV-2 antigens, generated many antibodies against ferritin as well as SARS-CoV-2.
“The DNA nanoparticle itself is immunogenically silent,” Lingwood says. “If you use a protein-based platform, you get equally high titer antibody responses to the platform and to the antigen of interest, and that can complicate repeated usage of that platform because you’ll develop high affinity immune memory against it.”
Reducing these off-target effects could also help scientists reach the goal of developing a vaccine that would induce broadly neutralizing antibodies to any variant of SARS-CoV-2, or even to all sarbecoviruses, the subgenus of virus that includes SARS-CoV-2 as well as the viruses that cause SARS and MERS.
To that end, the researchers are now exploring whether a DNA scaffold with many different viral antigens attached could induce broadly neutralizing antibodies against SARS-CoV-2 and related viruses.
The research was primarily funded by the National Institutes of Health, the National Science Foundation, and the Fast Grants program.

© Credit: The Bathe Lab
Tumors constantly shed DNA from dying cells, which briefly circulates in the patient’s bloodstream before it is quickly broken down. Many companies have created blood tests that can pick out this tumor DNA, potentially helping doctors diagnose or monitor cancer or choose a treatment.
The amount of tumor DNA circulating at any given time, however, is extremely small, so it has been challenging to develop tests sensitive enough to pick up that tiny signal. A team of researchers from MIT and the Broad Institute of MIT and Harvard has now come up with a way to significantly boost that signal, by temporarily slowing the clearance of tumor DNA circulating in the bloodstream.
The researchers developed two different types of injectable molecules that they call “priming agents,” which can transiently interfere with the body’s ability to remove circulating tumor DNA from the bloodstream. In a study of mice, they showed that these agents could boost DNA levels enough that the percentage of detectable early-stage lung metastases leapt from less than 10 percent to above 75 percent.
This approach could enable not only earlier diagnosis of cancer, but also more sensitive detection of tumor mutations that could be used to guide treatment. It could also help improve detection of cancer recurrence.
“You can give one of these agents an hour before the blood draw, and it makes things visible that previously wouldn’t have been. The implication is that we should be able to give everybody who’s doing liquid biopsies, for any purpose, more molecules to work with,” says Sangeeta Bhatia, the John and Dorothy Wilson Professor of Health Sciences and Technology and of Electrical Engineering and Computer Science at MIT, and a member of MIT’s Koch Institute for Integrative Cancer Research and the Institute for Medical Engineering and Science.
Bhatia is one of the senior authors of the new study, along with J. Christopher Love, the Raymond A. and Helen E. St. Laurent Professor of Chemical Engineering at MIT and a member of the Koch Institute and the Ragon Institute of MGH, MIT, and Harvard and Viktor Adalsteinsson, director of the Gerstner Center for Cancer Diagnostics at the Broad Institute.
Carmen Martin-Alonso PhD ’23, MIT and Broad Institute postdoc Shervin Tabrizi, and Broad Institute scientist Kan Xiong are the lead authors of the paper, which appears today in Science.
Better biopsies
Liquid biopsies, which enable detection of small quantities of DNA in blood samples, are now used in many cancer patients to identify mutations that could help guide treatment. With greater sensitivity, however, these tests could become useful for far more patients. Most efforts to improve the sensitivity of liquid biopsies have focused on developing new sequencing technologies to use after the blood is drawn.
While brainstorming ways to make liquid biopsies more informative, Bhatia, Love, Adalsteinsson, and their trainees came up with the idea of trying to increase the amount of DNA in a patient’s bloodstream before the sample is taken.
“A tumor is always creating new cell-free DNA, and that’s the signal that we’re attempting to detect in the blood draw. Existing liquid biopsy technologies, however, are limited by the amount of material you collect in the tube of blood,” Love says. “Where this work intercedes is thinking about how to inject something beforehand that would help boost or enhance the amount of signal that is available to collect in the same small sample.”
The body uses two primary strategies to remove circulating DNA from the bloodstream. Enzymes called DNases circulate in the blood and break down DNA that they encounter, while immune cells known as macrophages take up cell-free DNA as blood is filtered through the liver.
The researchers decided to target each of these processes separately. To prevent DNases from breaking down DNA, they designed a monoclonal antibody that binds to circulating DNA and protects it from the enzymes.
“Antibodies are well-established biopharmaceutical modalities, and they’re safe in a number of different disease contexts, including cancer and autoimmune treatments,” Love says. “The idea was, could we use this kind of antibody to help shield the DNA temporarily from degradation by the nucleases that are in circulation? And by doing so, we shift the balance to where the tumor is generating DNA slightly faster than is being degraded, increasing the concentration in a blood draw.”
The other priming agent they developed is a nanoparticle designed to block macrophages from taking up cell-free DNA. These cells have a well-known tendency to eat up synthetic nanoparticles.
“DNA is a biological nanoparticle, and it made sense that immune cells in the liver were probably taking this up just like they do synthetic nanoparticles. And if that were the case, which it turned out to be, then we could use a safe dummy nanoparticle to distract those immune cells and leave the circulating DNA alone so that it could be at a higher concentration,” Bhatia says.
Earlier tumor detection
The researchers tested their priming agents in mice that received transplants of cancer cells that tend to form tumors in the lungs. Two weeks after the cells were transplanted, the researchers showed that these priming agents could boost the amount of circulating tumor DNA recovered in a blood sample by up to 60-fold.
Once the blood sample is taken, it can be run through the same kinds of sequencing tests now used on liquid biopsy samples. These tests can pick out tumor DNA, including specific sequences used to determine the type of tumor and potentially what kinds of treatments would work best.
Early detection of cancer is another promising application for these priming agents. The researchers found that when mice were given the nanoparticle priming agent before blood was drawn, it allowed them to detect circulating tumor DNA in blood of 75 percent of the mice with low cancer burden, while none were detectable without this boost.
“One of the greatest hurdles for cancer liquid biopsy testing has been the scarcity of circulating tumor DNA in a blood sample,” Adalsteinsson says. “It’s thus been encouraging to see the magnitude of the effect we’ve been able to achieve so far and to envision what impact this could have for patients.”
After either of the priming agents are injected, it takes an hour or two for the DNA levels to increase in the bloodstream, and then they return to normal within about 24 hours.
“The ability to get peak activity of these agents within a couple of hours, followed by their rapid clearance, means that someone could go into a doctor’s office, receive an agent like this, and then give their blood for the test itself, all within one visit,” Love says. “This feature bodes well for the potential to translate this concept into clinical use.”
The researchers have launched a company called Amplifyer Bio that plans to further develop the technology, in hopes of advancing to clinical trials.
“A tube of blood is a much more accessible diagnostic than colonoscopy screening or even mammography,” Bhatia says. “Ultimately, if these tools really are predictive, then we should be able to get many more patients into the system who could benefit from cancer interception or better therapy.”
The research was funded by the Koch Institute Support (core) Grant from the National Cancer Institute, the Marble Center for Cancer Nanomedicine, the Gerstner Family Foundation, the Ludwig Center at MIT, the Koch Institute Frontier Research Program via the Casey and Family Foundation, and the Bridge Project, a partnership between the Koch Institute and the Dana-Farber/Harvard Cancer Center.

© Image: MIT News; iStock
Using a virus-like delivery particle made from DNA, researchers from MIT and the Ragon Institute of MGH, MIT, and Harvard have created a vaccine that can induce a strong antibody response against SARS-CoV-2.
The vaccine, which has been tested in mice, consists of a DNA scaffold that carries many copies of a viral antigen. This type of vaccine, known as a particulate vaccine, mimics the structure of a virus. Most previous work on particulate vaccines has relied on protein scaffolds, but the proteins used in those vaccines tend to generate an unnecessary immune response that can distract the immune system from the target.
In the mouse study, the researchers found that the DNA scaffold does not induce an immune response, allowing the immune system to focus its antibody response on the target antigen.
“DNA, we found in this work, does not elicit antibodies that may distract away from the protein of interest,” says Mark Bathe, an MIT professor of biological engineering. “What you can imagine is that your B cells and immune system are being fully trained by that target antigen, and that’s what you want — for your immune system to be laser-focused on the antigen of interest.”
This approach, which strongly stimulates B cells (the cells that produce antibodies), could make it easier to develop vaccines against viruses that have been difficult to target, including HIV and influenza, as well as SARS-CoV-2, the researchers say. Unlike T cells, which are stimulated by other types of vaccines, these B cells can persist for decades, offering long-term protection.
“We’re interested in exploring whether we can teach the immune system to deliver higher levels of immunity against pathogens that resist conventional vaccine approaches, like flu, HIV, and SARS-CoV-2,” says Daniel Lingwood, an associate professor at Harvard Medical School and a principal investigator at the Ragon Institute. “This idea of decoupling the response against the target antigen from the platform itself is a potentially powerful immunological trick that one can now bring to bear to help those immunological targeting decisions move in a direction that is more focused.”
Bathe, Lingwood, and Aaron Schmidt, an associate professor at Harvard Medical School and principal investigator at the Ragon Institute, are the senior authors of the paper, which appears today in Nature Communications. The paper’s lead authors are Eike-Christian Wamhoff, a former MIT postdoc; Larance Ronsard, a Ragon Institute postdoc; Jared Feldman, a former Harvard University graduate student; Grant Knappe, an MIT graduate student; and Blake Hauser, a former Harvard graduate student.
Mimicking viruses
Particulate vaccines usually consist of a protein nanoparticle, similar in structure to a virus, that can carry many copies of a viral antigen. This high density of antigens can lead to a stronger immune response than traditional vaccines because the body sees it as similar to an actual virus. Particulate vaccines have been developed for a handful of pathogens, including hepatitis B and human papillomavirus, and a particulate vaccine for SARS-CoV-2 has been approved for use in South Korea.
These vaccines are especially good at activating B cells, which produce antibodies specific to the vaccine antigen.
“Particulate vaccines are of great interest for many in immunology because they give you robust humoral immunity, which is antibody-based immunity, which is differentiated from the T-cell-based immunity that the mRNA vaccines seem to elicit more strongly,” Bathe says.
A potential drawback to this kind of vaccine, however, is that the proteins used for the scaffold often stimulate the body to produce antibodies targeting the scaffold. This can distract the immune system and prevent it from launching as robust a response as one would like, Bathe says.
“To neutralize the SARS-CoV-2 virus, you want to have a vaccine that generates antibodies toward the receptor binding domain portion of the virus’ spike protein,” he says. “When you display that on a protein-based particle, what happens is your immune system recognizes not only that receptor binding domain protein, but all the other proteins that are irrelevant to the immune response you’re trying to elicit.”
Another potential drawback is that if the same person receives more than one vaccine carried by the same protein scaffold, for example, SARS-CoV-2 and then influenza, their immune system would likely respond right away to the protein scaffold, having already been primed to react to it. This could weaken the immune response to the antigen carried by the second vaccine.
“If you want to apply that protein-based particle to immunize against a different virus like influenza, then your immune system can be addicted to the underlying protein scaffold that it’s already seen and developed an immune response toward,” Bathe says. “That can hypothetically diminish the quality of your antibody response for the actual antigen of interest.”
As an alternative, Bathe’s lab has been developing scaffolds made using DNA origami, a method that offers precise control over the structure of synthetic DNA and allows researchers to attach a variety of molecules, such as viral antigens, at specific locations.
In a 2020 study, Bathe and Darrell Irvine, an MIT professor of biological engineering and of materials science and engineering, showed that a DNA scaffold carrying 30 copies of an HIV antigen could generate a strong antibody response in B cells grown in the lab. This type of structure is optimal for activating B cells because it closely mimics the structure of nano-sized viruses, which display many copies of viral proteins in their surfaces.
“This approach builds off of a fundamental principle in B-cell antigen recognition, which is that if you have an arrayed display of the antigen, that promotes B-cell responses and gives better quantity and quality of antibody output,” Lingwood says.
“Immunologically silent”
In the new study, the researchers swapped in an antigen consisting of the receptor binding protein of the spike protein from the original strain of SARS-CoV-2. When they gave the vaccine to mice, they found that the mice generated high levels of antibodies to the spike protein but did not generate any to the DNA scaffold.
In contrast, a vaccine based on a scaffold protein called ferritin, coated with SARS-CoV-2 antigens, generated many antibodies against ferritin as well as SARS-CoV-2.
“The DNA nanoparticle itself is immunogenically silent,” Lingwood says. “If you use a protein-based platform, you get equally high titer antibody responses to the platform and to the antigen of interest, and that can complicate repeated usage of that platform because you’ll develop high affinity immune memory against it.”
Reducing these off-target effects could also help scientists reach the goal of developing a vaccine that would induce broadly neutralizing antibodies to any variant of SARS-CoV-2, or even to all sarbecoviruses, the subgenus of virus that includes SARS-CoV-2 as well as the viruses that cause SARS and MERS.
To that end, the researchers are now exploring whether a DNA scaffold with many different viral antigens attached could induce broadly neutralizing antibodies against SARS-CoV-2 and related viruses.
The research was primarily funded by the National Institutes of Health, the National Science Foundation, and the Fast Grants program.

© Credit: The Bathe Lab
Tumors constantly shed DNA from dying cells, which briefly circulates in the patient’s bloodstream before it is quickly broken down. Many companies have created blood tests that can pick out this tumor DNA, potentially helping doctors diagnose or monitor cancer or choose a treatment.
The amount of tumor DNA circulating at any given time, however, is extremely small, so it has been challenging to develop tests sensitive enough to pick up that tiny signal. A team of researchers from MIT and the Broad Institute of MIT and Harvard has now come up with a way to significantly boost that signal, by temporarily slowing the clearance of tumor DNA circulating in the bloodstream.
The researchers developed two different types of injectable molecules that they call “priming agents,” which can transiently interfere with the body’s ability to remove circulating tumor DNA from the bloodstream. In a study of mice, they showed that these agents could boost DNA levels enough that the percentage of detectable early-stage lung metastases leapt from less than 10 percent to above 75 percent.
This approach could enable not only earlier diagnosis of cancer, but also more sensitive detection of tumor mutations that could be used to guide treatment. It could also help improve detection of cancer recurrence.
“You can give one of these agents an hour before the blood draw, and it makes things visible that previously wouldn’t have been. The implication is that we should be able to give everybody who’s doing liquid biopsies, for any purpose, more molecules to work with,” says Sangeeta Bhatia, the John and Dorothy Wilson Professor of Health Sciences and Technology and of Electrical Engineering and Computer Science at MIT, and a member of MIT’s Koch Institute for Integrative Cancer Research and the Institute for Medical Engineering and Science.
Bhatia is one of the senior authors of the new study, along with J. Christopher Love, the Raymond A. and Helen E. St. Laurent Professor of Chemical Engineering at MIT and a member of the Koch Institute and the Ragon Institute of MGH, MIT, and Harvard and Viktor Adalsteinsson, director of the Gerstner Center for Cancer Diagnostics at the Broad Institute.
Carmen Martin-Alonso PhD ’23, MIT and Broad Institute postdoc Shervin Tabrizi, and Broad Institute scientist Kan Xiong are the lead authors of the paper, which appears today in Science.
Better biopsies
Liquid biopsies, which enable detection of small quantities of DNA in blood samples, are now used in many cancer patients to identify mutations that could help guide treatment. With greater sensitivity, however, these tests could become useful for far more patients. Most efforts to improve the sensitivity of liquid biopsies have focused on developing new sequencing technologies to use after the blood is drawn.
While brainstorming ways to make liquid biopsies more informative, Bhatia, Love, Adalsteinsson, and their trainees came up with the idea of trying to increase the amount of DNA in a patient’s bloodstream before the sample is taken.
“A tumor is always creating new cell-free DNA, and that’s the signal that we’re attempting to detect in the blood draw. Existing liquid biopsy technologies, however, are limited by the amount of material you collect in the tube of blood,” Love says. “Where this work intercedes is thinking about how to inject something beforehand that would help boost or enhance the amount of signal that is available to collect in the same small sample.”
The body uses two primary strategies to remove circulating DNA from the bloodstream. Enzymes called DNases circulate in the blood and break down DNA that they encounter, while immune cells known as macrophages take up cell-free DNA as blood is filtered through the liver.
The researchers decided to target each of these processes separately. To prevent DNases from breaking down DNA, they designed a monoclonal antibody that binds to circulating DNA and protects it from the enzymes.
“Antibodies are well-established biopharmaceutical modalities, and they’re safe in a number of different disease contexts, including cancer and autoimmune treatments,” Love says. “The idea was, could we use this kind of antibody to help shield the DNA temporarily from degradation by the nucleases that are in circulation? And by doing so, we shift the balance to where the tumor is generating DNA slightly faster than is being degraded, increasing the concentration in a blood draw.”
The other priming agent they developed is a nanoparticle designed to block macrophages from taking up cell-free DNA. These cells have a well-known tendency to eat up synthetic nanoparticles.
“DNA is a biological nanoparticle, and it made sense that immune cells in the liver were probably taking this up just like they do synthetic nanoparticles. And if that were the case, which it turned out to be, then we could use a safe dummy nanoparticle to distract those immune cells and leave the circulating DNA alone so that it could be at a higher concentration,” Bhatia says.
Earlier tumor detection
The researchers tested their priming agents in mice that received transplants of cancer cells that tend to form tumors in the lungs. Two weeks after the cells were transplanted, the researchers showed that these priming agents could boost the amount of circulating tumor DNA recovered in a blood sample by up to 60-fold.
Once the blood sample is taken, it can be run through the same kinds of sequencing tests now used on liquid biopsy samples. These tests can pick out tumor DNA, including specific sequences used to determine the type of tumor and potentially what kinds of treatments would work best.
Early detection of cancer is another promising application for these priming agents. The researchers found that when mice were given the nanoparticle priming agent before blood was drawn, it allowed them to detect circulating tumor DNA in blood of 75 percent of the mice with low cancer burden, while none were detectable without this boost.
“One of the greatest hurdles for cancer liquid biopsy testing has been the scarcity of circulating tumor DNA in a blood sample,” Adalsteinsson says. “It’s thus been encouraging to see the magnitude of the effect we’ve been able to achieve so far and to envision what impact this could have for patients.”
After either of the priming agents are injected, it takes an hour or two for the DNA levels to increase in the bloodstream, and then they return to normal within about 24 hours.
“The ability to get peak activity of these agents within a couple of hours, followed by their rapid clearance, means that someone could go into a doctor’s office, receive an agent like this, and then give their blood for the test itself, all within one visit,” Love says. “This feature bodes well for the potential to translate this concept into clinical use.”
The researchers have launched a company called Amplifyer Bio that plans to further develop the technology, in hopes of advancing to clinical trials.
“A tube of blood is a much more accessible diagnostic than colonoscopy screening or even mammography,” Bhatia says. “Ultimately, if these tools really are predictive, then we should be able to get many more patients into the system who could benefit from cancer interception or better therapy.”
The research was funded by the Koch Institute Support (core) Grant from the National Cancer Institute, the Marble Center for Cancer Nanomedicine, the Gerstner Family Foundation, the Ludwig Center at MIT, the Koch Institute Frontier Research Program via the Casey and Family Foundation, and the Bridge Project, a partnership between the Koch Institute and the Dana-Farber/Harvard Cancer Center.

© Image: MIT News; iStock
Using a virus-like delivery particle made from DNA, researchers from MIT and the Ragon Institute of MGH, MIT, and Harvard have created a vaccine that can induce a strong antibody response against SARS-CoV-2.
The vaccine, which has been tested in mice, consists of a DNA scaffold that carries many copies of a viral antigen. This type of vaccine, known as a particulate vaccine, mimics the structure of a virus. Most previous work on particulate vaccines has relied on protein scaffolds, but the proteins used in those vaccines tend to generate an unnecessary immune response that can distract the immune system from the target.
In the mouse study, the researchers found that the DNA scaffold does not induce an immune response, allowing the immune system to focus its antibody response on the target antigen.
“DNA, we found in this work, does not elicit antibodies that may distract away from the protein of interest,” says Mark Bathe, an MIT professor of biological engineering. “What you can imagine is that your B cells and immune system are being fully trained by that target antigen, and that’s what you want — for your immune system to be laser-focused on the antigen of interest.”
This approach, which strongly stimulates B cells (the cells that produce antibodies), could make it easier to develop vaccines against viruses that have been difficult to target, including HIV and influenza, as well as SARS-CoV-2, the researchers say. Unlike T cells, which are stimulated by other types of vaccines, these B cells can persist for decades, offering long-term protection.
“We’re interested in exploring whether we can teach the immune system to deliver higher levels of immunity against pathogens that resist conventional vaccine approaches, like flu, HIV, and SARS-CoV-2,” says Daniel Lingwood, an associate professor at Harvard Medical School and a principal investigator at the Ragon Institute. “This idea of decoupling the response against the target antigen from the platform itself is a potentially powerful immunological trick that one can now bring to bear to help those immunological targeting decisions move in a direction that is more focused.”
Bathe, Lingwood, and Aaron Schmidt, an associate professor at Harvard Medical School and principal investigator at the Ragon Institute, are the senior authors of the paper, which appears today in Nature Communications. The paper’s lead authors are Eike-Christian Wamhoff, a former MIT postdoc; Larance Ronsard, a Ragon Institute postdoc; Jared Feldman, a former Harvard University graduate student; Grant Knappe, an MIT graduate student; and Blake Hauser, a former Harvard graduate student.
Mimicking viruses
Particulate vaccines usually consist of a protein nanoparticle, similar in structure to a virus, that can carry many copies of a viral antigen. This high density of antigens can lead to a stronger immune response than traditional vaccines because the body sees it as similar to an actual virus. Particulate vaccines have been developed for a handful of pathogens, including hepatitis B and human papillomavirus, and a particulate vaccine for SARS-CoV-2 has been approved for use in South Korea.
These vaccines are especially good at activating B cells, which produce antibodies specific to the vaccine antigen.
“Particulate vaccines are of great interest for many in immunology because they give you robust humoral immunity, which is antibody-based immunity, which is differentiated from the T-cell-based immunity that the mRNA vaccines seem to elicit more strongly,” Bathe says.
A potential drawback to this kind of vaccine, however, is that the proteins used for the scaffold often stimulate the body to produce antibodies targeting the scaffold. This can distract the immune system and prevent it from launching as robust a response as one would like, Bathe says.
“To neutralize the SARS-CoV-2 virus, you want to have a vaccine that generates antibodies toward the receptor binding domain portion of the virus’ spike protein,” he says. “When you display that on a protein-based particle, what happens is your immune system recognizes not only that receptor binding domain protein, but all the other proteins that are irrelevant to the immune response you’re trying to elicit.”
Another potential drawback is that if the same person receives more than one vaccine carried by the same protein scaffold, for example, SARS-CoV-2 and then influenza, their immune system would likely respond right away to the protein scaffold, having already been primed to react to it. This could weaken the immune response to the antigen carried by the second vaccine.
“If you want to apply that protein-based particle to immunize against a different virus like influenza, then your immune system can be addicted to the underlying protein scaffold that it’s already seen and developed an immune response toward,” Bathe says. “That can hypothetically diminish the quality of your antibody response for the actual antigen of interest.”
As an alternative, Bathe’s lab has been developing scaffolds made using DNA origami, a method that offers precise control over the structure of synthetic DNA and allows researchers to attach a variety of molecules, such as viral antigens, at specific locations.
In a 2020 study, Bathe and Darrell Irvine, an MIT professor of biological engineering and of materials science and engineering, showed that a DNA scaffold carrying 30 copies of an HIV antigen could generate a strong antibody response in B cells grown in the lab. This type of structure is optimal for activating B cells because it closely mimics the structure of nano-sized viruses, which display many copies of viral proteins in their surfaces.
“This approach builds off of a fundamental principle in B-cell antigen recognition, which is that if you have an arrayed display of the antigen, that promotes B-cell responses and gives better quantity and quality of antibody output,” Lingwood says.
“Immunologically silent”
In the new study, the researchers swapped in an antigen consisting of the receptor binding protein of the spike protein from the original strain of SARS-CoV-2. When they gave the vaccine to mice, they found that the mice generated high levels of antibodies to the spike protein but did not generate any to the DNA scaffold.
In contrast, a vaccine based on a scaffold protein called ferritin, coated with SARS-CoV-2 antigens, generated many antibodies against ferritin as well as SARS-CoV-2.
“The DNA nanoparticle itself is immunogenically silent,” Lingwood says. “If you use a protein-based platform, you get equally high titer antibody responses to the platform and to the antigen of interest, and that can complicate repeated usage of that platform because you’ll develop high affinity immune memory against it.”
Reducing these off-target effects could also help scientists reach the goal of developing a vaccine that would induce broadly neutralizing antibodies to any variant of SARS-CoV-2, or even to all sarbecoviruses, the subgenus of virus that includes SARS-CoV-2 as well as the viruses that cause SARS and MERS.
To that end, the researchers are now exploring whether a DNA scaffold with many different viral antigens attached could induce broadly neutralizing antibodies against SARS-CoV-2 and related viruses.
The research was primarily funded by the National Institutes of Health, the National Science Foundation, and the Fast Grants program.

© Credit: The Bathe Lab
Tumors constantly shed DNA from dying cells, which briefly circulates in the patient’s bloodstream before it is quickly broken down. Many companies have created blood tests that can pick out this tumor DNA, potentially helping doctors diagnose or monitor cancer or choose a treatment.
The amount of tumor DNA circulating at any given time, however, is extremely small, so it has been challenging to develop tests sensitive enough to pick up that tiny signal. A team of researchers from MIT and the Broad Institute of MIT and Harvard has now come up with a way to significantly boost that signal, by temporarily slowing the clearance of tumor DNA circulating in the bloodstream.
The researchers developed two different types of injectable molecules that they call “priming agents,” which can transiently interfere with the body’s ability to remove circulating tumor DNA from the bloodstream. In a study of mice, they showed that these agents could boost DNA levels enough that the percentage of detectable early-stage lung metastases leapt from less than 10 percent to above 75 percent.
This approach could enable not only earlier diagnosis of cancer, but also more sensitive detection of tumor mutations that could be used to guide treatment. It could also help improve detection of cancer recurrence.
“You can give one of these agents an hour before the blood draw, and it makes things visible that previously wouldn’t have been. The implication is that we should be able to give everybody who’s doing liquid biopsies, for any purpose, more molecules to work with,” says Sangeeta Bhatia, the John and Dorothy Wilson Professor of Health Sciences and Technology and of Electrical Engineering and Computer Science at MIT, and a member of MIT’s Koch Institute for Integrative Cancer Research and the Institute for Medical Engineering and Science.
Bhatia is one of the senior authors of the new study, along with J. Christopher Love, the Raymond A. and Helen E. St. Laurent Professor of Chemical Engineering at MIT and a member of the Koch Institute and the Ragon Institute of MGH, MIT, and Harvard and Viktor Adalsteinsson, director of the Gerstner Center for Cancer Diagnostics at the Broad Institute.
Carmen Martin-Alonso PhD ’23, MIT and Broad Institute postdoc Shervin Tabrizi, and Broad Institute scientist Kan Xiong are the lead authors of the paper, which appears today in Science.
Better biopsies
Liquid biopsies, which enable detection of small quantities of DNA in blood samples, are now used in many cancer patients to identify mutations that could help guide treatment. With greater sensitivity, however, these tests could become useful for far more patients. Most efforts to improve the sensitivity of liquid biopsies have focused on developing new sequencing technologies to use after the blood is drawn.
While brainstorming ways to make liquid biopsies more informative, Bhatia, Love, Adalsteinsson, and their trainees came up with the idea of trying to increase the amount of DNA in a patient’s bloodstream before the sample is taken.
“A tumor is always creating new cell-free DNA, and that’s the signal that we’re attempting to detect in the blood draw. Existing liquid biopsy technologies, however, are limited by the amount of material you collect in the tube of blood,” Love says. “Where this work intercedes is thinking about how to inject something beforehand that would help boost or enhance the amount of signal that is available to collect in the same small sample.”
The body uses two primary strategies to remove circulating DNA from the bloodstream. Enzymes called DNases circulate in the blood and break down DNA that they encounter, while immune cells known as macrophages take up cell-free DNA as blood is filtered through the liver.
The researchers decided to target each of these processes separately. To prevent DNases from breaking down DNA, they designed a monoclonal antibody that binds to circulating DNA and protects it from the enzymes.
“Antibodies are well-established biopharmaceutical modalities, and they’re safe in a number of different disease contexts, including cancer and autoimmune treatments,” Love says. “The idea was, could we use this kind of antibody to help shield the DNA temporarily from degradation by the nucleases that are in circulation? And by doing so, we shift the balance to where the tumor is generating DNA slightly faster than is being degraded, increasing the concentration in a blood draw.”
The other priming agent they developed is a nanoparticle designed to block macrophages from taking up cell-free DNA. These cells have a well-known tendency to eat up synthetic nanoparticles.
“DNA is a biological nanoparticle, and it made sense that immune cells in the liver were probably taking this up just like they do synthetic nanoparticles. And if that were the case, which it turned out to be, then we could use a safe dummy nanoparticle to distract those immune cells and leave the circulating DNA alone so that it could be at a higher concentration,” Bhatia says.
Earlier tumor detection
The researchers tested their priming agents in mice that received transplants of cancer cells that tend to form tumors in the lungs. Two weeks after the cells were transplanted, the researchers showed that these priming agents could boost the amount of circulating tumor DNA recovered in a blood sample by up to 60-fold.
Once the blood sample is taken, it can be run through the same kinds of sequencing tests now used on liquid biopsy samples. These tests can pick out tumor DNA, including specific sequences used to determine the type of tumor and potentially what kinds of treatments would work best.
Early detection of cancer is another promising application for these priming agents. The researchers found that when mice were given the nanoparticle priming agent before blood was drawn, it allowed them to detect circulating tumor DNA in blood of 75 percent of the mice with low cancer burden, while none were detectable without this boost.
“One of the greatest hurdles for cancer liquid biopsy testing has been the scarcity of circulating tumor DNA in a blood sample,” Adalsteinsson says. “It’s thus been encouraging to see the magnitude of the effect we’ve been able to achieve so far and to envision what impact this could have for patients.”
After either of the priming agents are injected, it takes an hour or two for the DNA levels to increase in the bloodstream, and then they return to normal within about 24 hours.
“The ability to get peak activity of these agents within a couple of hours, followed by their rapid clearance, means that someone could go into a doctor’s office, receive an agent like this, and then give their blood for the test itself, all within one visit,” Love says. “This feature bodes well for the potential to translate this concept into clinical use.”
The researchers have launched a company called Amplifyer Bio that plans to further develop the technology, in hopes of advancing to clinical trials.
“A tube of blood is a much more accessible diagnostic than colonoscopy screening or even mammography,” Bhatia says. “Ultimately, if these tools really are predictive, then we should be able to get many more patients into the system who could benefit from cancer interception or better therapy.”
The research was funded by the Koch Institute Support (core) Grant from the National Cancer Institute, the Marble Center for Cancer Nanomedicine, the Gerstner Family Foundation, the Ludwig Center at MIT, the Koch Institute Frontier Research Program via the Casey and Family Foundation, and the Bridge Project, a partnership between the Koch Institute and the Dana-Farber/Harvard Cancer Center.

© Image: MIT News; iStock
Flat screen TVs that incorporate quantum dots are now commercially available, but it has been more difficult to create arrays of their elongated cousins, quantum rods, for commercial devices. Quantum rods can control both the polarization and color of light, to generate 3D images for virtual reality devices.
Using scaffolds made of folded DNA, MIT engineers have come up with a new way to precisely assemble arrays of quantum rods. By depositing quantum rods onto a DNA scaffold in a highly controlled way, the researchers can regulate their orientation, which is a key factor in determining the polarization of light emitted by the array. This makes it easier to add depth and dimensionality to a virtual scene.
“One of the challenges with quantum rods is: How do you align them all at the nanoscale so they’re all pointing in the same direction?” says Mark Bathe, an MIT professor of biological engineering and the senior author of the new study. “When they’re all pointing in the same direction on a 2D surface, then they all have the same properties of how they interact with light and control its polarization.”
MIT postdocs Chi Chen and Xin Luo are the lead authors of the paper, which appears today in Science Advances. Robert Macfarlane, an associate professor of materials science and engineering; Alexander Kaplan PhD ’23; and Moungi Bawendi, the Lester Wolfe Professor of Chemistry, are also authors of the study.
Nanoscale structures
Over the past 15 years, Bathe and others have led in the design and fabrication of nanoscale structures made of DNA, also known as DNA origami. DNA, a highly stable and programmable molecule, is an ideal building material for tiny structures that could be used for a variety of applications, including delivering drugs, acting as biosensors, or forming scaffolds for light-harvesting materials.
Bathe’s lab has developed computational methods that allow researchers to simply enter a target nanoscale shape they want to create, and the program will calculate the sequences of DNA that will self-assemble into the right shape. They also developed scalable fabrication methods that incorporate quantum dots into these DNA-based materials.
In a 2022 paper, Bathe and Chen showed that they could use DNA to scaffold quantum dots in precise positions using scalable biological fabrication. Building on that work, they teamed up with Macfarlane’s lab to tackle the challenge of arranging quantum rods into 2D arrays, which is more difficult because the rods need to be aligned in the same direction.
Existing approaches that create aligned arrays of quantum rods using mechanical rubbing with a fabric or an electric field to sweep the rods into one direction have had only limited success. This is because high-efficiency light-emission requires the rods to be kept at least 10 nanometers from each other, so that they won’t “quench,” or suppress, their neighbors’ light-emitting activity.
To achieve that, the researchers devised a way to attach quantum rods to diamond-shaped DNA origami structures, which can be built at the right size to maintain that distance. These DNA structures are then attached to a surface, where they fit together like puzzle pieces.
“The quantum rods sit on the origami in the same direction, so now you have patterned all these quantum rods through self-assembly on 2D surfaces, and you can do that over the micron scale needed for different applications like microLEDs,” Bathe says. “You can orient them in specific directions that are controllable and keep them well-separated because the origamis are packed and naturally fit together, as puzzle pieces would.”
Assembling the puzzle
As the first step in getting this approach to work, the researchers had to come up with a way to attach DNA strands to the quantum rods. To do that, Chen developed a process that involves emulsifying DNA into a mixture with the quantum rods, then rapidly dehydrating the mixture, which allows the DNA molecules to form a dense layer on the surface of the rods.
This process takes only a few minutes, much faster than any existing method for attaching DNA to nanoscale particles, which may be key to enabling commercial applications.
“The unique aspect of this method lies in its near-universal applicability to any water-loving ligand with affinity to the nanoparticle surface, allowing them to be instantly pushed onto the surface of the nanoscale particles. By harnessing this method, we achieved a significant reduction in manufacturing time from several days to just a few minutes,” Chen says.
These DNA strands then act like Velcro, helping the quantum rods stick to a DNA origami template, which forms a thin film that coats a silicate surface. This thin film of DNA is first formed via self-assembly by joining neighboring DNA templates together via overhanging strands of DNA along their edges.
The researchers now hope to create wafer-scale surfaces with etched patterns, which could allow them to scale their design to device-scale arrangements of quantum rods for numerous applications, beyond only microLEDs or augmented reality/virtual reality.
“The method that we describe in this paper is great because it provides good spatial and orientational control of how the quantum rods are positioned. The next steps are going to be making arrays that are more hierarchical, with programmed structure at many different length scales. The ability to control the sizes, shapes, and placement of these quantum rod arrays is a gateway to all sorts of different electronics applications,” Macfarlane says.
“DNA is particularly attractive as a manufacturing material because it can be biologically produced, which is both scalable and sustainable, in line with the emerging U.S. bioeconomy. Translating this work toward commercial devices by solving several remaining bottlenecks, including switching to environmentally safe quantum rods, is what we’re focused on next,” Bathe adds.
The research was funded by the Office of Naval Research, the National Science Foundation, the Army Research Office, the Department of Energy, and the National Institute of Environmental Health Sciences.

© Image: Dr. Xin Luo, Bathe BioNanoLab

How much thought do you give to where you keep your bits? Every day we produce more data, including emails, texts, photos, and social media posts. Though much of this content is forgettable, every day we implicitly decide not to get rid of that data. We keep it somewhere, be it in on a phone, on a computer’s hard drive, or in the cloud, where it is eventually archived, in most cases on magnetic tape. Consider further the many varied devices and sensors now streaming data onto the Web, and the cars, airplanes, and other vehicles that store trip data for later use. All those billions of things on the Internet of Things produce data, and all that information also needs to be stored somewhere.
Data is piling up exponentially, and the rate of information production is increasing faster than the storage density of tape, which will only be able to keep up with the deluge of data for a few more years. The research firm Gartner predicts that by 2030, the shortfall in enterprise storage capacity alone could amount to nearly two-thirds of demand, or about 20 million petabytes. If we continue down our current path, in coming decades we would need not only exponentially more magnetic tape, disk drives, and flash memory, but exponentially more factories to produce these storage media, and exponentially more data centers and warehouses to store them. Even if this is technically feasible, it’s economically implausible.
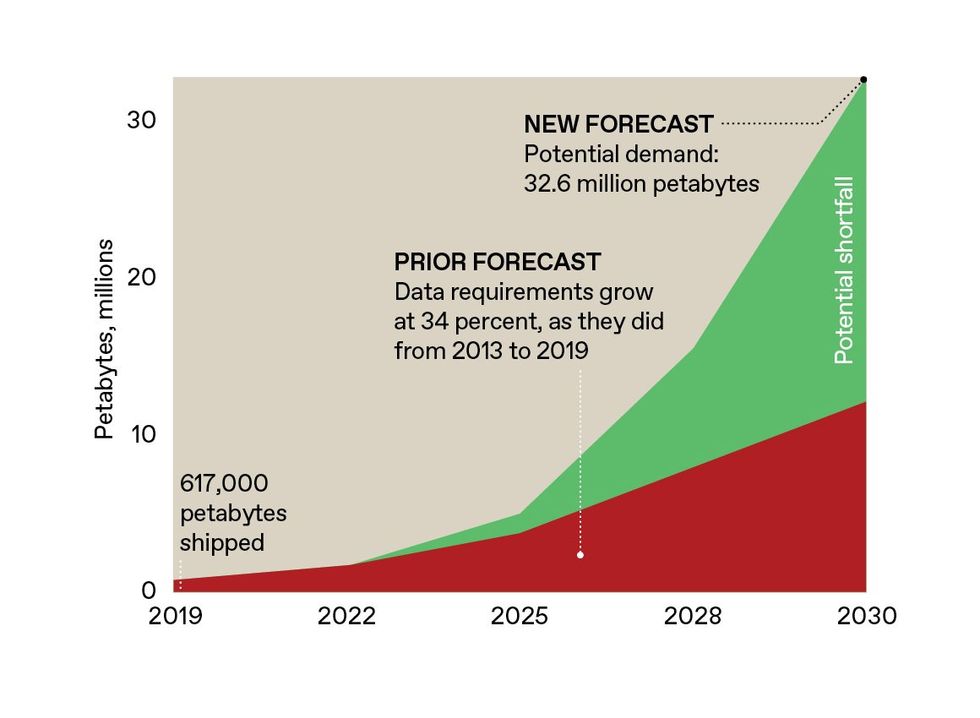 Prior projections for data storage requirements estimated a global need for about 12 million petabytes of capacity by 2030. The research firm Gartner recently issued new projections, raising that estimate by 20 million petabytes. The world is not on track to produce enough of today’s storage technologies to fill that gap.SOURCE: GARTNER
Prior projections for data storage requirements estimated a global need for about 12 million petabytes of capacity by 2030. The research firm Gartner recently issued new projections, raising that estimate by 20 million petabytes. The world is not on track to produce enough of today’s storage technologies to fill that gap.SOURCE: GARTNER
Fortunately, we have access to an information storage technology that is cheap, readily available, and stable at room temperature for millennia: DNA, the material of genes. In a few years your hard drive may be full of such squishy stuff.
Storing information in DNA is not a complicated concept. Decades ago, humans learned to sequence and synthesize DNA—that is, to read and write it. Each position in a single strand of DNA consists of one of four nucleic acids, known as bases and represented as A, T, G, and C. In principle, each position in the DNA strand could be used to store two bits (A could represent 00, T could be 01, and so on), but in practice, information is generally stored at an effective one bit—a 0 or a 1—per base.
Moreover, DNA exceeds by many times the storage density of magnetic tape or solid-state media. It has been calculated that all the information on the Internet—which one estimate puts at about 120 zettabytes—could be stored in a volume of DNA about the size of a sugar cube, or approximately a cubic centimeter. Achieving that density is theoretically possible, but we could get by with a much lower storage density. An effective storage density of “one Internet per 1,000 cubic meters” would still result in something considerably smaller than a single data center housing tape today.
 In 2018, researchers built this first prototype of a machine that could write, store, and read data with DNA.MICROSOFT RESEARCH
In 2018, researchers built this first prototype of a machine that could write, store, and read data with DNA.MICROSOFT RESEARCH
Most examples of DNA data storage to date rely on chemically synthesizing short stretches of DNA, up to 200 or so bases. Standard chemical synthesis methods are adequate for demonstration projects, and perhaps early commercial efforts, that store modest amounts of music, images, text, and video, up to perhaps hundreds of gigabytes. However, as the technology matures, we will need to switch from chemical synthesis to a much more elegant, scalable, and sustainable solution: a semiconductor chip that uses enzymes to write these sequences.
After the data has been written into the DNA, the molecule must be kept safe somewhere. Published examples include drying small spots of DNA on glass or paper, encasing the DNA in sugar or silica particles, or just putting it in a test tube. Reading can be accomplished with any number of commercial sequencing technologies.
Organizations around the world are already taking the first steps toward building a DNA drive that can both write and read DNA data. I’ve participated in this effort via a collaboration between Microsoft and the Molecular Information Systems Lab of the Paul G. Allen School of Computer Science and Engineering at the University of Washington. We’ve made considerable progress already, and we can see the way forward.
First, let’s look at the current state of storage. As mentioned, magnetic tape storage has a scaling problem. Making matters worse, tape degrades quickly compared to the time scale on which we want to store information. To last longer than a decade, tape must be carefully stored at cool temperatures and low humidity, which typically means the continuous use of energy for air conditioning. And even when stored carefully, tape needs to be replaced periodically, so we need more tape not just for all the new data but to replace the tape storing the old data.
To be sure, the storage density of magnetic tape has been increasing for decades, a trend that will help keep our heads above the data flood for a while longer. But current practices are building fragility into the storage ecosystem. Backward compatibility is often guaranteed for only a generation or two of the hardware used to read that media, which could be just a few years, requiring the active maintenance of aging hardware or ongoing data migration. So all the data we have already stored digitally is at risk of being lost to technological obsolescence.
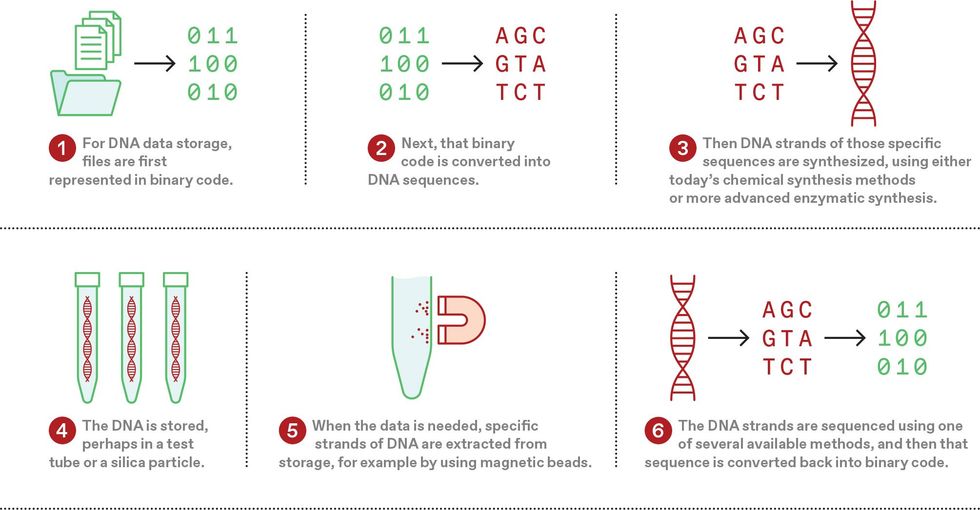
The discussion thus far has assumed that we’ll want to keep all the data we produce, and that we’ll pay to do so. We should entertain the counterhypothesis: that we will instead engage in systematic forgetting on a global scale. This voluntary amnesia might be accomplished by not collecting as much data about the world or by not saving all the data we collect, perhaps only keeping derivative calculations and conclusions. Or maybe not every person or organization will have the same access to storage. If it becomes a limited resource, data storage could become a strategic technology that enables a company, or a country, to capture and process all the data it desires, while competitors suffer a storage deficit. But as yet, there’s no sign that producers of data are willing to lose any of it.
If we are to avoid either accidental or intentional forgetting, we need to come up with a fundamentally different solution for storing data, one with the potential for exponential improvements far beyond those expected for tape. DNA is by far the most sophisticated, stable, and dense information-storage technology humans have ever come across or invented. Readable genomic DNA has been recovered after having been frozen in the tundra for 2 million years. DNA is an intrinsic part of life on this planet. As best we can tell, nucleic acid–based genetic information storage has persisted on Earth for at least 3 billion years, giving it an unassailable advantage as a backward- and forward-compatible data storage medium.
To date, humans have learned to sequence and synthesize short pieces of single-stranded DNA (ssDNA). However, in naturally occurring genomes, DNA is usually in the form of long, double-stranded DNA (dsDNA). This dsDNA is composed of two complementary sequences bound into a structure that resembles a twisting ladder, where sugar backbones form the side rails, and the paired bases—A with T, and G with C—form the steps of the ladder. Due to this structure, dsDNA is generally more robust than ssDNA.
Reading and writing DNA are both noisy molecular processes. To enable resiliency in the presence of this noise, digital information is encoded using an algorithm that introduces redundancy and distributes information across many bases. Current algorithms encode information at a physical density of 1 bit per 60 atoms (a pair of bases and the sugar backbones to which they’re attached).
 Edmon de Haro
Edmon de Haro
Synthesizing and sequencing DNA has become critical to the global economy, to human health, and to understanding how organisms and ecosystems are changing around us. And we’re likely to only get better at it over time. Indeed, both the cost and the per-instrument throughput of writing and reading DNA have been improving exponentially for decades, roughly keeping up with Moore’s Law.
In biology labs around the world, it’s now common practice to order chemically synthesized ssDNA from a commercial provider; these molecules are delivered in lengths of up to several hundred bases. It is also common to sequence DNA molecules that are up to thousands of bases in length. In other words, we already convert digital information to and from DNA, but generally using only sequences that make sense in terms of biology.
For DNA data storage, though, we will have to write arbitrary sequences that are much longer, probably thousands to tens of thousands of bases. We’ll do that by adapting the naturally occurring biological process and fusing it with semiconductor technology to create high-density input and output devices.
There is global interest in creating a DNA drive. The members of the DNA Data Storage Alliance, founded in 2020, come from universities, companies of all sizes, and government labs from around the world. Funding agencies in the United States, Europe, and Asia are investing in the technology stack required to field commercially relevant devices. Potential customers as diverse as film studios, the U.S. National Archives, and Boeing have expressed interest in long-term data storage in DNA.
Archival storage might be the first market to emerge, given that it involves writing once with only infrequent reading, and yet also demands stability over many decades, if not centuries. Storing information in DNA for that time span is easily achievable. The challenging part is learning how to get the information into, and back out of, the molecule in an economically viable way.
The first soup-to-nuts automated prototype capable of writing, storing, and reading DNA was built by my Microsoft and University of Washington colleagues in 2018. The prototype integrated standard plumbing and chemistry to write the DNA, with a sequencer from the company Oxford Nanopore Technologies to read the DNA. This single-channel device, which occupied a tabletop, had a throughput of 5 bytes over approximately 21 hours, with all but 40 minutes of that time consumed in writing “HELLO” into the DNA. It was a start.
For a DNA drive to compete with today’s archival tape drives, it must be able to write about 2 gigabits per second, which at demonstrated DNA data storage densities is about 2 billion bases per second. To put that in context, I estimate that the total global market for synthetic DNA today is no more than about 10 terabases per year, which is the equivalent of about 300,000 bases per second over a year. The entire DNA synthesis industry would need to grow by approximately 4 orders of magnitude just to compete with a single tape drive. Keeping up with the total global demand for storage would require another 8 orders of magnitude of improvement by 2030.
Exponential growth in silicon-based technology is how we wound up producing so much data. Similar exponential growth will be fundamental in the transition to DNA storage.
But humans have done this kind of scaling up before. Exponential growth in silicon-based technology is how we wound up producing so much data. Similar exponential growth will be fundamental in the transition to DNA storage.
My work with colleagues at the University of Washington and Microsoft has yielded many promising results. This collaboration has made progress on error-tolerant encoding of DNA, writing information into DNA sequences, stably storing that DNA, and recovering the information by reading the DNA. The team has also explored the economic, environmental, and architectural advantages of DNA data storage compared to alternatives.
One of our goals was to build a semiconductor chip to enable high-density, high-throughput DNA synthesis. That chip, which we completed in 2021, demonstrated that it is possible to digitally control electrochemical processes in millions of 650-nanometer-diameter wells. While the chip itself was a technological step forward, the chemical synthesis we used on that chip had a few drawbacks, despite being the industry standard. The main problem is that it employs a volatile, corrosive, and toxic organic solvent (acetonitrile), which no engineer wants anywhere near the electronics of a working data center.
Moreover, based on a sustainability analysis of a theoretical DNA data center performed my colleagues at Microsoft, I conclude that the volume of acetonitrile required for just one large data center, never mind many large data centers, would become logistically and economically prohibitive. To be sure, each data center could be equipped with a recycling facility to reuse the solvent, but that would be costly.
Fortunately, there is a different emerging technology for constructing DNA that does not require such solvents, but instead uses a benign salt solution. Companies like DNA Script and Molecular Assemblies are commercializing automated systems that use enzymes to synthesize DNA. These techniques are replacing traditional chemical DNA synthesis for some applications in the biotechnology industry. The current generation of systems use either simple plumbing or light to control synthesis reactions. But it’s difficult to envision how they can be scaled to achieve a high enough throughput to enable a DNA data-storage device operating at even a fraction of 2 gigabases per second.
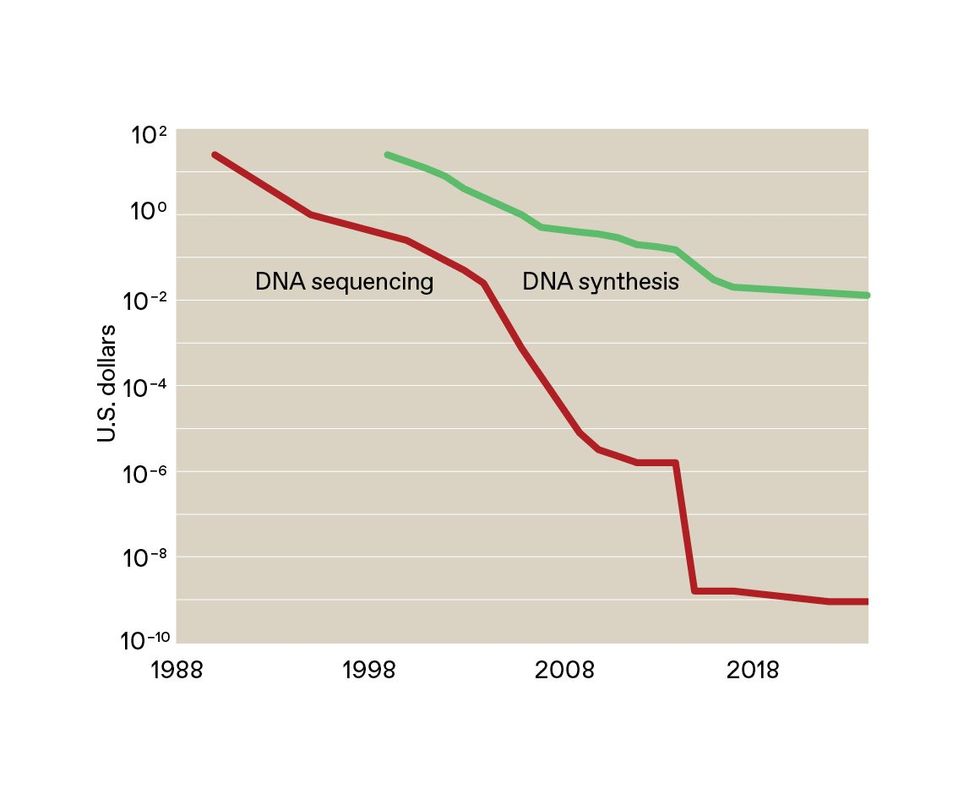 The price for sequencing DNA has plummeted from $25 per base in 1990 to less than a millionth of a cent in 2024. The cost of synthesizing long pieces of double-stranded DNA is also declining, but synthesis needs to become much cheaper for DNA data storage to really take off.SOURCE: ROB CARLSON
The price for sequencing DNA has plummeted from $25 per base in 1990 to less than a millionth of a cent in 2024. The cost of synthesizing long pieces of double-stranded DNA is also declining, but synthesis needs to become much cheaper for DNA data storage to really take off.SOURCE: ROB CARLSON
Still, the enzymes inside these systems are important pieces of the DNA drive puzzle. Like DNA data storage, the idea of using enzymes to write DNA is not new, but commercial enzymatic synthesis became feasible only in the last couple of years. Most such processes use an enzyme called terminal deoxynucleotidyl transferase, or TdT. Whereas most enzymes that operate on DNA use one strand as a template to fill in the other strand, TdT can add arbitrary bases to single-stranded DNA.
Naturally occurring TdT is not a great enzyme for synthesis, because it incorporates the four bases with four different efficiencies, and it’s hard to control. Efforts over the past decade have focused on modifying the TdT and building it into a system in which the enzyme can be better controlled.
Notably, those modifications to TdT were made possible by prior decades of improvement in reading and writing DNA, and the new modified enzymes are now contributing to further improvements in writing, and thus modifying, genes and genomes. This phenomenon is the same type of feedback that drove decades of exponential improvement in the semiconductor industry, in which companies used more capable silicon chips to design the next generation of silicon chips. Because that feedback continues apace in both arenas, it won’t be long before we can combine the two technologies into one functional device: a semiconductor chip that converts digital signals into chemical states (for example, changes in pH), and an enzymatic system that responds to those chemical states by adding specific, individual bases to build a strand of synthetic DNA.
The University of Washington and Microsoft team, collaborating with the enzymatic synthesis company Ansa Biotechnologies, recently took the first step toward this device. Using our high-density chip, we successfully demonstrated electrochemical control of single-base enzymatic additions. The project is now paused while the team evaluates possible next steps.Nevertheless, even if this effort is not resumed, someone will make the technology work. The path is relatively clear; building a commercially relevant DNA drive is simply a matter of time and money.
Eventually, the technology for DNA storage will completely alter the economics of reading and writing all kinds of genetic information. Even if the performance bar is set far below that of a tape drive, any commercial operation based on reading and writing data into DNA will have a throughput many times that of today’s DNA synthesis industry, with a vanishingly small cost per base.
At the same time, advances in DNA synthesis for DNA storage will increase access to DNA for other uses, notably in the biotechnology industry, and will thereby expand capabilities to reprogram life. Somewhere down the road, when a DNA drive achieves a throughput of 2 gigabases per second (or 120 gigabases per minute), this box could synthesize the equivalent of about 20 complete human genomes per minute. And when humans combine our improving knowledge of how to construct a genome with access to effectively free synthetic DNA, we will enter a very different world.
The conversations we have today about biosecurity, who has access to DNA synthesis, and whether this technology can be controlled are barely scratching the surface of what is to come. We’ll be able to design microbes to produce chemicals and drugs, as well as plants that can fend off pests or sequester minerals from the environment, such as arsenic, carbon, or gold. At 2 gigabases per second, constructing biological countermeasures against novel pathogens will take a matter of minutes. But so too will constructing the genomes of novel pathogens. Indeed, this flow of information back and forth between the digital and the biological will mean that every security concern from the world of IT will also be introduced into the world of biology. We will have to be vigilant about these possibilities.
We are just beginning to learn how to build and program systems that integrate digital logic and biochemistry. The future will be built not from DNA as we find it, but from DNA as we will write it.
This article appears in the March 2024 print issue.

AMD GPUOpen - Graphics and game developer resources
The second post in this series on mesh shaders covers best practices for writing mesh and amplification shaders, as well as how to use the AMD Radeon™ Developer Tool Suite to profile and optimize mesh shaders.
The post Optimization and best practices – Mesh shaders on RDNA™ graphics cards appeared first on AMD GPUOpen.
Using a virus-like delivery particle made from DNA, researchers from MIT and the Ragon Institute of MGH, MIT, and Harvard have created a vaccine that can induce a strong antibody response against SARS-CoV-2.
The vaccine, which has been tested in mice, consists of a DNA scaffold that carries many copies of a viral antigen. This type of vaccine, known as a particulate vaccine, mimics the structure of a virus. Most previous work on particulate vaccines has relied on protein scaffolds, but the proteins used in those vaccines tend to generate an unnecessary immune response that can distract the immune system from the target.
In the mouse study, the researchers found that the DNA scaffold does not induce an immune response, allowing the immune system to focus its antibody response on the target antigen.
“DNA, we found in this work, does not elicit antibodies that may distract away from the protein of interest,” says Mark Bathe, an MIT professor of biological engineering. “What you can imagine is that your B cells and immune system are being fully trained by that target antigen, and that’s what you want — for your immune system to be laser-focused on the antigen of interest.”
This approach, which strongly stimulates B cells (the cells that produce antibodies), could make it easier to develop vaccines against viruses that have been difficult to target, including HIV and influenza, as well as SARS-CoV-2, the researchers say. Unlike T cells, which are stimulated by other types of vaccines, these B cells can persist for decades, offering long-term protection.
“We’re interested in exploring whether we can teach the immune system to deliver higher levels of immunity against pathogens that resist conventional vaccine approaches, like flu, HIV, and SARS-CoV-2,” says Daniel Lingwood, an associate professor at Harvard Medical School and a principal investigator at the Ragon Institute. “This idea of decoupling the response against the target antigen from the platform itself is a potentially powerful immunological trick that one can now bring to bear to help those immunological targeting decisions move in a direction that is more focused.”
Bathe, Lingwood, and Aaron Schmidt, an associate professor at Harvard Medical School and principal investigator at the Ragon Institute, are the senior authors of the paper, which appears today in Nature Communications. The paper’s lead authors are Eike-Christian Wamhoff, a former MIT postdoc; Larance Ronsard, a Ragon Institute postdoc; Jared Feldman, a former Harvard University graduate student; Grant Knappe, an MIT graduate student; and Blake Hauser, a former Harvard graduate student.
Mimicking viruses
Particulate vaccines usually consist of a protein nanoparticle, similar in structure to a virus, that can carry many copies of a viral antigen. This high density of antigens can lead to a stronger immune response than traditional vaccines because the body sees it as similar to an actual virus. Particulate vaccines have been developed for a handful of pathogens, including hepatitis B and human papillomavirus, and a particulate vaccine for SARS-CoV-2 has been approved for use in South Korea.
These vaccines are especially good at activating B cells, which produce antibodies specific to the vaccine antigen.
“Particulate vaccines are of great interest for many in immunology because they give you robust humoral immunity, which is antibody-based immunity, which is differentiated from the T-cell-based immunity that the mRNA vaccines seem to elicit more strongly,” Bathe says.
A potential drawback to this kind of vaccine, however, is that the proteins used for the scaffold often stimulate the body to produce antibodies targeting the scaffold. This can distract the immune system and prevent it from launching as robust a response as one would like, Bathe says.
“To neutralize the SARS-CoV-2 virus, you want to have a vaccine that generates antibodies toward the receptor binding domain portion of the virus’ spike protein,” he says. “When you display that on a protein-based particle, what happens is your immune system recognizes not only that receptor binding domain protein, but all the other proteins that are irrelevant to the immune response you’re trying to elicit.”
Another potential drawback is that if the same person receives more than one vaccine carried by the same protein scaffold, for example, SARS-CoV-2 and then influenza, their immune system would likely respond right away to the protein scaffold, having already been primed to react to it. This could weaken the immune response to the antigen carried by the second vaccine.
“If you want to apply that protein-based particle to immunize against a different virus like influenza, then your immune system can be addicted to the underlying protein scaffold that it’s already seen and developed an immune response toward,” Bathe says. “That can hypothetically diminish the quality of your antibody response for the actual antigen of interest.”
As an alternative, Bathe’s lab has been developing scaffolds made using DNA origami, a method that offers precise control over the structure of synthetic DNA and allows researchers to attach a variety of molecules, such as viral antigens, at specific locations.
In a 2020 study, Bathe and Darrell Irvine, an MIT professor of biological engineering and of materials science and engineering, showed that a DNA scaffold carrying 30 copies of an HIV antigen could generate a strong antibody response in B cells grown in the lab. This type of structure is optimal for activating B cells because it closely mimics the structure of nano-sized viruses, which display many copies of viral proteins in their surfaces.
“This approach builds off of a fundamental principle in B-cell antigen recognition, which is that if you have an arrayed display of the antigen, that promotes B-cell responses and gives better quantity and quality of antibody output,” Lingwood says.
“Immunologically silent”
In the new study, the researchers swapped in an antigen consisting of the receptor binding protein of the spike protein from the original strain of SARS-CoV-2. When they gave the vaccine to mice, they found that the mice generated high levels of antibodies to the spike protein but did not generate any to the DNA scaffold.
In contrast, a vaccine based on a scaffold protein called ferritin, coated with SARS-CoV-2 antigens, generated many antibodies against ferritin as well as SARS-CoV-2.
“The DNA nanoparticle itself is immunogenically silent,” Lingwood says. “If you use a protein-based platform, you get equally high titer antibody responses to the platform and to the antigen of interest, and that can complicate repeated usage of that platform because you’ll develop high affinity immune memory against it.”
Reducing these off-target effects could also help scientists reach the goal of developing a vaccine that would induce broadly neutralizing antibodies to any variant of SARS-CoV-2, or even to all sarbecoviruses, the subgenus of virus that includes SARS-CoV-2 as well as the viruses that cause SARS and MERS.
To that end, the researchers are now exploring whether a DNA scaffold with many different viral antigens attached could induce broadly neutralizing antibodies against SARS-CoV-2 and related viruses.
The research was primarily funded by the National Institutes of Health, the National Science Foundation, and the Fast Grants program.

© Credit: The Bathe Lab
Tumors constantly shed DNA from dying cells, which briefly circulates in the patient’s bloodstream before it is quickly broken down. Many companies have created blood tests that can pick out this tumor DNA, potentially helping doctors diagnose or monitor cancer or choose a treatment.
The amount of tumor DNA circulating at any given time, however, is extremely small, so it has been challenging to develop tests sensitive enough to pick up that tiny signal. A team of researchers from MIT and the Broad Institute of MIT and Harvard has now come up with a way to significantly boost that signal, by temporarily slowing the clearance of tumor DNA circulating in the bloodstream.
The researchers developed two different types of injectable molecules that they call “priming agents,” which can transiently interfere with the body’s ability to remove circulating tumor DNA from the bloodstream. In a study of mice, they showed that these agents could boost DNA levels enough that the percentage of detectable early-stage lung metastases leapt from less than 10 percent to above 75 percent.
This approach could enable not only earlier diagnosis of cancer, but also more sensitive detection of tumor mutations that could be used to guide treatment. It could also help improve detection of cancer recurrence.
“You can give one of these agents an hour before the blood draw, and it makes things visible that previously wouldn’t have been. The implication is that we should be able to give everybody who’s doing liquid biopsies, for any purpose, more molecules to work with,” says Sangeeta Bhatia, the John and Dorothy Wilson Professor of Health Sciences and Technology and of Electrical Engineering and Computer Science at MIT, and a member of MIT’s Koch Institute for Integrative Cancer Research and the Institute for Medical Engineering and Science.
Bhatia is one of the senior authors of the new study, along with J. Christopher Love, the Raymond A. and Helen E. St. Laurent Professor of Chemical Engineering at MIT and a member of the Koch Institute and the Ragon Institute of MGH, MIT, and Harvard and Viktor Adalsteinsson, director of the Gerstner Center for Cancer Diagnostics at the Broad Institute.
Carmen Martin-Alonso PhD ’23, MIT and Broad Institute postdoc Shervin Tabrizi, and Broad Institute scientist Kan Xiong are the lead authors of the paper, which appears today in Science.
Better biopsies
Liquid biopsies, which enable detection of small quantities of DNA in blood samples, are now used in many cancer patients to identify mutations that could help guide treatment. With greater sensitivity, however, these tests could become useful for far more patients. Most efforts to improve the sensitivity of liquid biopsies have focused on developing new sequencing technologies to use after the blood is drawn.
While brainstorming ways to make liquid biopsies more informative, Bhatia, Love, Adalsteinsson, and their trainees came up with the idea of trying to increase the amount of DNA in a patient’s bloodstream before the sample is taken.
“A tumor is always creating new cell-free DNA, and that’s the signal that we’re attempting to detect in the blood draw. Existing liquid biopsy technologies, however, are limited by the amount of material you collect in the tube of blood,” Love says. “Where this work intercedes is thinking about how to inject something beforehand that would help boost or enhance the amount of signal that is available to collect in the same small sample.”
The body uses two primary strategies to remove circulating DNA from the bloodstream. Enzymes called DNases circulate in the blood and break down DNA that they encounter, while immune cells known as macrophages take up cell-free DNA as blood is filtered through the liver.
The researchers decided to target each of these processes separately. To prevent DNases from breaking down DNA, they designed a monoclonal antibody that binds to circulating DNA and protects it from the enzymes.
“Antibodies are well-established biopharmaceutical modalities, and they’re safe in a number of different disease contexts, including cancer and autoimmune treatments,” Love says. “The idea was, could we use this kind of antibody to help shield the DNA temporarily from degradation by the nucleases that are in circulation? And by doing so, we shift the balance to where the tumor is generating DNA slightly faster than is being degraded, increasing the concentration in a blood draw.”
The other priming agent they developed is a nanoparticle designed to block macrophages from taking up cell-free DNA. These cells have a well-known tendency to eat up synthetic nanoparticles.
“DNA is a biological nanoparticle, and it made sense that immune cells in the liver were probably taking this up just like they do synthetic nanoparticles. And if that were the case, which it turned out to be, then we could use a safe dummy nanoparticle to distract those immune cells and leave the circulating DNA alone so that it could be at a higher concentration,” Bhatia says.
Earlier tumor detection
The researchers tested their priming agents in mice that received transplants of cancer cells that tend to form tumors in the lungs. Two weeks after the cells were transplanted, the researchers showed that these priming agents could boost the amount of circulating tumor DNA recovered in a blood sample by up to 60-fold.
Once the blood sample is taken, it can be run through the same kinds of sequencing tests now used on liquid biopsy samples. These tests can pick out tumor DNA, including specific sequences used to determine the type of tumor and potentially what kinds of treatments would work best.
Early detection of cancer is another promising application for these priming agents. The researchers found that when mice were given the nanoparticle priming agent before blood was drawn, it allowed them to detect circulating tumor DNA in blood of 75 percent of the mice with low cancer burden, while none were detectable without this boost.
“One of the greatest hurdles for cancer liquid biopsy testing has been the scarcity of circulating tumor DNA in a blood sample,” Adalsteinsson says. “It’s thus been encouraging to see the magnitude of the effect we’ve been able to achieve so far and to envision what impact this could have for patients.”
After either of the priming agents are injected, it takes an hour or two for the DNA levels to increase in the bloodstream, and then they return to normal within about 24 hours.
“The ability to get peak activity of these agents within a couple of hours, followed by their rapid clearance, means that someone could go into a doctor’s office, receive an agent like this, and then give their blood for the test itself, all within one visit,” Love says. “This feature bodes well for the potential to translate this concept into clinical use.”
The researchers have launched a company called Amplifyer Bio that plans to further develop the technology, in hopes of advancing to clinical trials.
“A tube of blood is a much more accessible diagnostic than colonoscopy screening or even mammography,” Bhatia says. “Ultimately, if these tools really are predictive, then we should be able to get many more patients into the system who could benefit from cancer interception or better therapy.”
The research was funded by the Koch Institute Support (core) Grant from the National Cancer Institute, the Marble Center for Cancer Nanomedicine, the Gerstner Family Foundation, the Ludwig Center at MIT, the Koch Institute Frontier Research Program via the Casey and Family Foundation, and the Bridge Project, a partnership between the Koch Institute and the Dana-Farber/Harvard Cancer Center.

© Image: MIT News; iStock
Flat screen TVs that incorporate quantum dots are now commercially available, but it has been more difficult to create arrays of their elongated cousins, quantum rods, for commercial devices. Quantum rods can control both the polarization and color of light, to generate 3D images for virtual reality devices.
Using scaffolds made of folded DNA, MIT engineers have come up with a new way to precisely assemble arrays of quantum rods. By depositing quantum rods onto a DNA scaffold in a highly controlled way, the researchers can regulate their orientation, which is a key factor in determining the polarization of light emitted by the array. This makes it easier to add depth and dimensionality to a virtual scene.
“One of the challenges with quantum rods is: How do you align them all at the nanoscale so they’re all pointing in the same direction?” says Mark Bathe, an MIT professor of biological engineering and the senior author of the new study. “When they’re all pointing in the same direction on a 2D surface, then they all have the same properties of how they interact with light and control its polarization.”
MIT postdocs Chi Chen and Xin Luo are the lead authors of the paper, which appears today in Science Advances. Robert Macfarlane, an associate professor of materials science and engineering; Alexander Kaplan PhD ’23; and Moungi Bawendi, the Lester Wolfe Professor of Chemistry, are also authors of the study.
Nanoscale structures
Over the past 15 years, Bathe and others have led in the design and fabrication of nanoscale structures made of DNA, also known as DNA origami. DNA, a highly stable and programmable molecule, is an ideal building material for tiny structures that could be used for a variety of applications, including delivering drugs, acting as biosensors, or forming scaffolds for light-harvesting materials.
Bathe’s lab has developed computational methods that allow researchers to simply enter a target nanoscale shape they want to create, and the program will calculate the sequences of DNA that will self-assemble into the right shape. They also developed scalable fabrication methods that incorporate quantum dots into these DNA-based materials.
In a 2022 paper, Bathe and Chen showed that they could use DNA to scaffold quantum dots in precise positions using scalable biological fabrication. Building on that work, they teamed up with Macfarlane’s lab to tackle the challenge of arranging quantum rods into 2D arrays, which is more difficult because the rods need to be aligned in the same direction.
Existing approaches that create aligned arrays of quantum rods using mechanical rubbing with a fabric or an electric field to sweep the rods into one direction have had only limited success. This is because high-efficiency light-emission requires the rods to be kept at least 10 nanometers from each other, so that they won’t “quench,” or suppress, their neighbors’ light-emitting activity.
To achieve that, the researchers devised a way to attach quantum rods to diamond-shaped DNA origami structures, which can be built at the right size to maintain that distance. These DNA structures are then attached to a surface, where they fit together like puzzle pieces.
“The quantum rods sit on the origami in the same direction, so now you have patterned all these quantum rods through self-assembly on 2D surfaces, and you can do that over the micron scale needed for different applications like microLEDs,” Bathe says. “You can orient them in specific directions that are controllable and keep them well-separated because the origamis are packed and naturally fit together, as puzzle pieces would.”
Assembling the puzzle
As the first step in getting this approach to work, the researchers had to come up with a way to attach DNA strands to the quantum rods. To do that, Chen developed a process that involves emulsifying DNA into a mixture with the quantum rods, then rapidly dehydrating the mixture, which allows the DNA molecules to form a dense layer on the surface of the rods.
This process takes only a few minutes, much faster than any existing method for attaching DNA to nanoscale particles, which may be key to enabling commercial applications.
“The unique aspect of this method lies in its near-universal applicability to any water-loving ligand with affinity to the nanoparticle surface, allowing them to be instantly pushed onto the surface of the nanoscale particles. By harnessing this method, we achieved a significant reduction in manufacturing time from several days to just a few minutes,” Chen says.
These DNA strands then act like Velcro, helping the quantum rods stick to a DNA origami template, which forms a thin film that coats a silicate surface. This thin film of DNA is first formed via self-assembly by joining neighboring DNA templates together via overhanging strands of DNA along their edges.
The researchers now hope to create wafer-scale surfaces with etched patterns, which could allow them to scale their design to device-scale arrangements of quantum rods for numerous applications, beyond only microLEDs or augmented reality/virtual reality.
“The method that we describe in this paper is great because it provides good spatial and orientational control of how the quantum rods are positioned. The next steps are going to be making arrays that are more hierarchical, with programmed structure at many different length scales. The ability to control the sizes, shapes, and placement of these quantum rod arrays is a gateway to all sorts of different electronics applications,” Macfarlane says.
“DNA is particularly attractive as a manufacturing material because it can be biologically produced, which is both scalable and sustainable, in line with the emerging U.S. bioeconomy. Translating this work toward commercial devices by solving several remaining bottlenecks, including switching to environmentally safe quantum rods, is what we’re focused on next,” Bathe adds.
The research was funded by the Office of Naval Research, the National Science Foundation, the Army Research Office, the Department of Energy, and the National Institute of Environmental Health Sciences.

© Image: Dr. Xin Luo, Bathe BioNanoLab